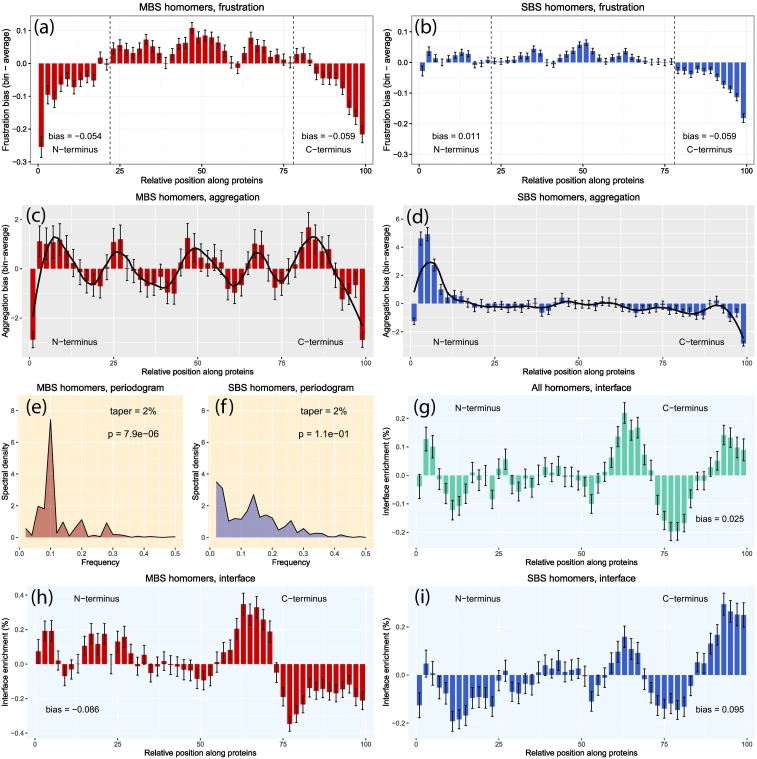
Fig. 6.
The sequence and structural signatures of chaperone interactions and complex assembly are different in single-domain MBS and SBS homomers. (A and B) Distribution of frustration bias along the protein sequences (lower is more frustrated). MBS homomers have significantly elevated levels of frustration at both termini of the sequence, but there is no difference between the two termini (p = 0.19, randomization test), while in SBS homomers, only the C terminus is frustrated (p < 0.0001, rand. test). (C and D) The distribution of aggregation propensity along the protein sequences. Bias was measured as the difference from the average of the proteins. In MBS homomers, the pattern is characterized by clear periodicity, although of modest magnitude, while in SBS homomers, it is characterized with N-terminal enrichment (p < 0.0001, rand. test). (E and F) The periodograms of aggregation bias indicate that the periodicity is highly significant in MBS homomers but not in SBS homomers (Fisher's G tests). (G, H and I) Distributions of interface enrichment in all BioLiP homomers (G), MBS homomers (H) and SBS homomers (I). MBS homomers show a depletion of interface residues at the C terminus (p = 0.0001, rand. test), while SBS homomers show an enrichment of interface residues (p = 0.0001, rand. test), indicating that cotranslational assembly influences the location of the interface only in SBS homomers. In the pooled data set of homomers (G), there is no significant C-terminal bias (p = 0.1, rand. test). On every plot, whiskers represent standard deviations.