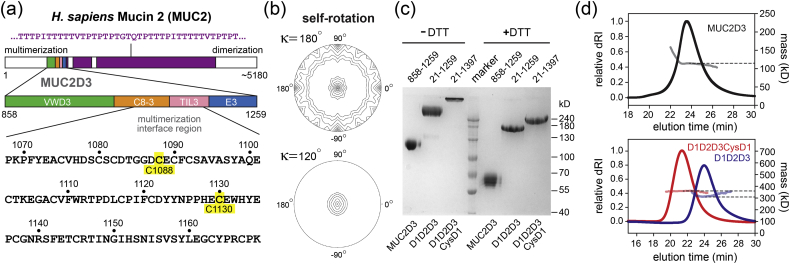
Fig. 1.
MUC2D3 organization. (a) Primary structural map of human MUC2 (Uniprot Q02817) shows the location of MUC2D3 within the full-length protein, the domain composition of MUC2D3, and the location of the intersubunit disulfide bonds (highlighted in yellow) in the amino acid sequence. A representative segment of the low-complexity (threonine-rich) sequence (purple) is displayed. (b) The κ = 180° (top) and κ = 120° (bottom) sections of the self-rotation function of the MUC2D3 diffraction data show that molecules in the crystal are related by a rotation of ~ 120° around an axis coincident with a crystallographic 2-fold symmetry axis. (c) SDS-PAGE (7.5% gel) of glycosylated MUC2D3 and longer MUC2 fragments without reduction by dithiothreitol (− DTT) and following reduction (+ DTT). Amino acid ranges for each fragment are given above the gel, and the corresponding domain designations are shown below. D1D2D3 spans MUC2 residues 21 to 1259; D1D2D3CysD1 spans residues 21 to 1397. (d) SEC-MALS analysis of MUC2D3 (above) and longer MUC2 fragments (below). The signals from the differential refractrometer (dRI) were normalized to yield peak values of 1 for MUC2D3 and D1D2D3CysD1, and the dRI signal of D1D2D3 was scaled by the same factor as D1D2D3CysD1. Dashed lines intersect the plots of molecular mass at the dRI peaks. The higher masses detected at the leading edge of the MUC2D3 peak are likely to be due to some non-covalent aggregation of dimers, rather than to a higher-order covalent assembly, because no hint of additional covalent multimers was seen by SDS-PAGE, even in overloaded gels. Calculated subunit masses (protein component only and including the carboxy-terminal His6 tag) are as follows: 45,700 Da for MUC2D3; 137,500 for D1D2D3, and 152,700 for D1D2D3CysD1.