Figure 1.
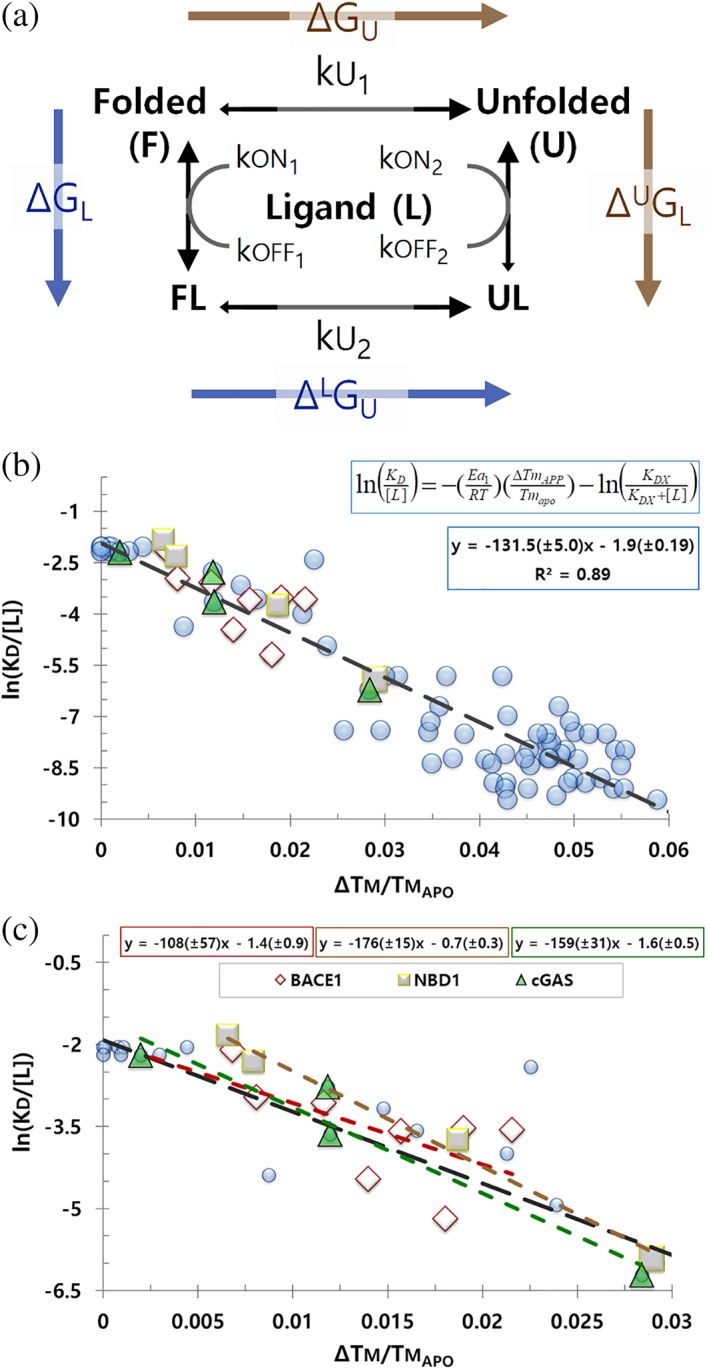
Model and ΔTm data for irreversible denaturation. (a) Irreversible unfolding model: apo folded (F), ligand‐bound folded (FL), apo unfolded (U), and ligand‐bound unfolded (UL). Ligand (L) has a significant preference for F over U; protein does not readily refold. Thermodynamic cycle in blue and gold. (b) v plot for irreversible unfolding. Proteins include the published values for BACE1 (diamonds),7 cGAS (squares),16 the NBD1 domain of CFTR (triangles),17 or other proteins available at Pfizer (circles). Fit and trendline for all proteins is shown; K D span ca 10 to 0.05 μM. (c) Individual fits and trendlines for BACE1, cGAS, and NBD1 with their ligands; the black line is the global trendline for all proteins from panel b. Protein and ligands are not identified if they are part of an active therapeutic research program at Pfizer. All data in these figures are available to download
