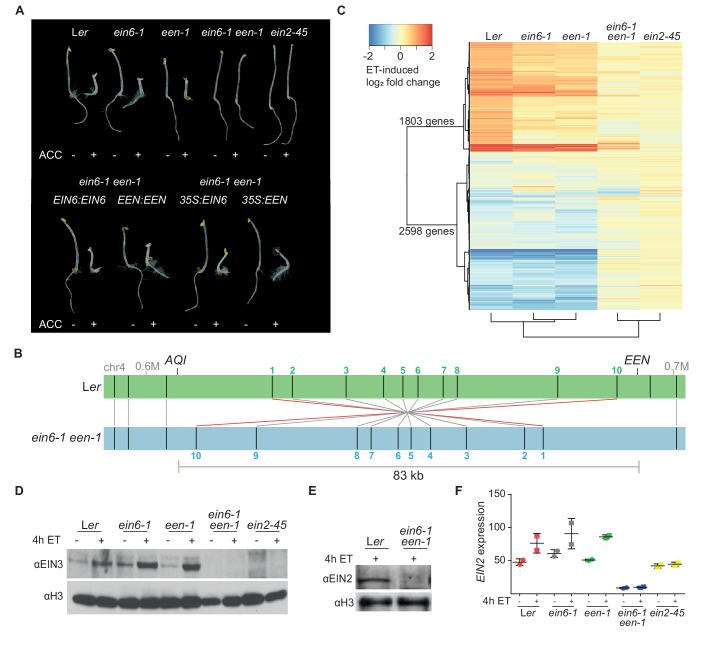
Figure 1. Mutations in two different genes are responsible for the ET-insensitivity in ein6-1 plants.
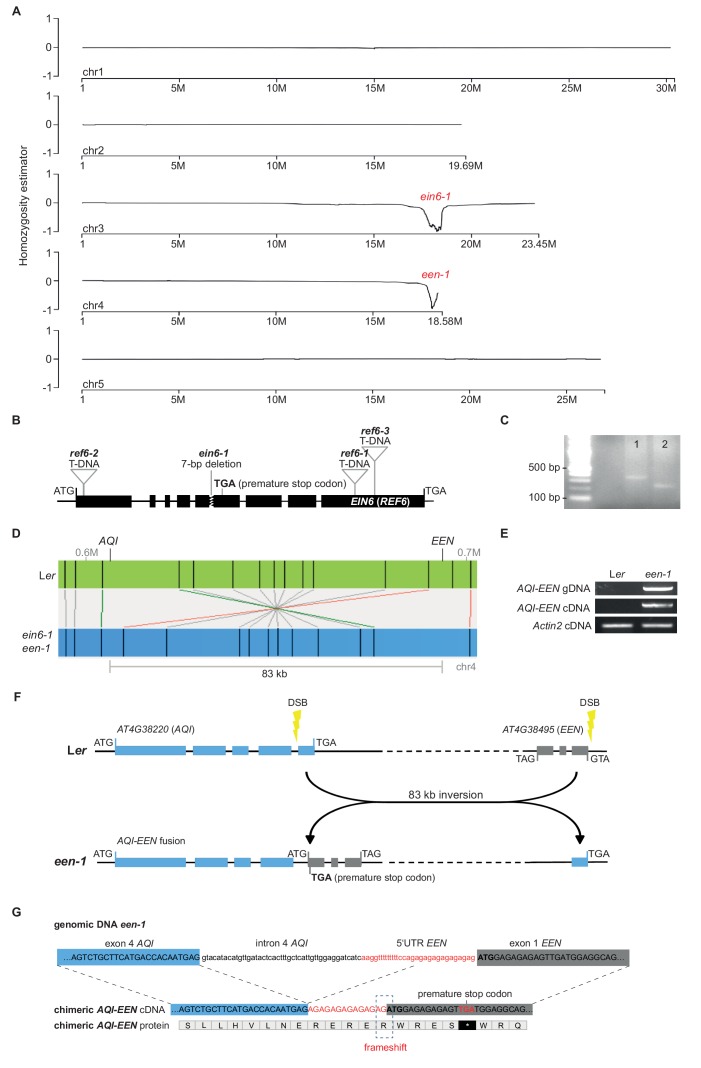
(A) Triple response phenotype of 3-day-old etiolated seedlings of Ler, ein6-1, een-1, ein6-1 een-1, ein2-45 (upper row) and of the indicated complementation lines, either driven by the respective native promoter or by the Cauliflower mosaic virus 35S promoter (35S) (lower row). Seedlings were grown on control LS media or LS media supplemented with 10 µM ACC. (B) Schematic illustration of a Bionano Genomics Irys optical map of the een-1 inversion region at the end of the fourth chromosome in ein6-1 een-1 (blue) aligned to an optical map of Ler (green). Original output is shown in Figure 1—figure supplement 1D. Nick sites are indicated as black lines within the respective optical map. Matching nick sites between the maps are indicated as gray lines. Numbered nick sites are used to better visualize the inversion event. The approximate position of EEN and AQI is indicated as well. (C) Heatmap visualizes the log2 fold change of expression in Ler, ein6-1, een-1, ein6-1 een-1 and ein2-45 seedlings in response to 4 hr of ethylene (ET) treatment detected by RNA-seq. Differentially expressed genes (DE genes) that are significantly induced (1803 genes) or repressed (2598 genes) after 4 hr of ET treatment in Ler seedlings are shown. Cluster dendogram below the heatmap indicate similarities between the tested genotypes. (D), (E), Western blot analysis of nuclear extracts of 3-day-old etiolated Ler, ein6-1, een-1, ein6-1 een-1 and ein2-45 seedlings that were either treated for 4 hr with hydrocarbon-free air (control) or ET gas. Antibodies against EIN3 and EIN2 were used to detect nuclear-localized EIN3 (D) and EIN2C (E), respectively. Amounts of histone H3 were detected with an anti-H3 antibody and served as a loading control. (F) EIN2 expression in response to 4 hr of ET treatment in the indicated genotypes using RNA-seq is shown. EIN2 transcripts can still be detected in ein2-45 mutants because they only harbor a point mutation in the C-terminus of EIN2 (Beaudoin et al., 2000). Expression is measured in TPM (Transcripts Per Kilobase Million) and results from two biological replicates are shown.