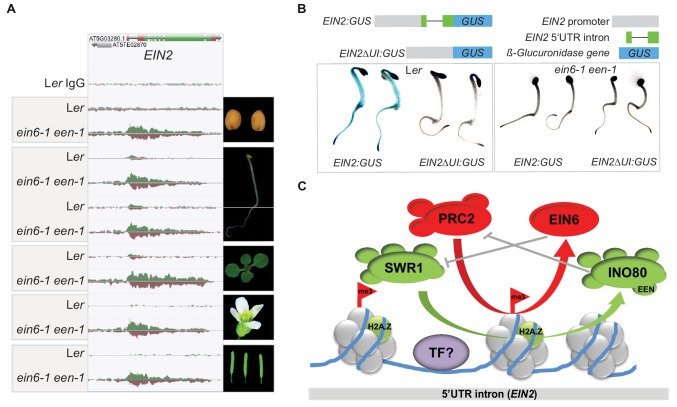
Figure 5. EIN6 (REF6)/INO80-mediated regulation occurs at the 5’UTR intron region of EIN2 during embryogenesis.
(A) Genome browser screenshot visualizes levels of H3K27me3 at the EIN2 gene in the indicated tissues of Ler and ein6-1 een-1 plants (dry seeds, roots and shoots of etiolated seedlings, de-etiolated whole-seedlings, flowers and siliques). H3K27me3 occupancy was identified with ChIP-seq. The tracks were normalized to the respective sequencing depth for each experiment to allow an accurate comparison between Ler and ein6-1 een-1 H3K27me3 profiles. Normalization was separately done for each chromatin feature. (B) Histochemical GUS staining of untreated three-day-old etiolated Ler and ein6-1 een-1 seedlings stably expressing either a EIN2 promoter GUS fusions with (EIN2:GUS) or without the 5’UTR intron (EIN2ΔI:GUS). Schematic illustration of the used EIN2:GUS constructs is shown as well. The gray box indicates the 2 kb promoter region of EIN2, the green boxes represent the 5’UTR and the black line indicates the 5’UTR intron. (C) Hypothetic model of the EIN6 (REF6)/INO80-mediated double safeguard mechanism at the 5’UTR intron of EIN2. Besides the demethylase function role of EIN6 (REF6) in antagonizing PRC2-mediated tri-methylation of H3K27, we speculate that EIN6 (REF6) also antagonizes SWR1-facillitated H2A.Z incorporation at EIN2. This scenario of a dual function holds true for the INO80 complex as well which counteracts SWR1 by removing H2A.Z, but also antagonizes H3K27 trimethylation through an unknown mechanism. The resulting low levels of H3K27me3 and H2A.Z possibly increase the accessibility of the enhancer element for binding of yet unknown transcription factors (TFs). When this EIN6 (REF6)/INO80-mediated regulation is functionally impaired, H3K27me3 and H2A.Z start to accumulate over the entire gene body of EIN2 mutually reinforcing their depositions.