Abstract
Modular plasmid architectures have shown to be a very useful resource to standardize, build, share, and compare biological parts and functional vectors, and are being applied in an increasing number of microorganisms. Here, we present a modular plasmid toolkit for Thermus thermophilus, a species considered as a workhorse for biotechnology and a model for high-temperature biology. Apart from integrating improved versions of already existing parts, we have characterized specific promoters and developed a thermosensor-based palette that restricts the expression to Thermus and, at the same time, controls protein expression in this organism in a temperature-dependent manner.
Introduction
Modular plasmid systems have been described for a number of taxons (see ref1 for a review). These systems represent clear improvements since they comprehend and enhance traditional toolkits commonly used to operate with the corresponding host species. Standardization of parts and components, as in other fields, is highly desirable as it has generally streamlined assembly and made observations easier to compare. Expectably, most of the modular plasmid systems have been developed for mesophilic bacteria; however, there are two modular systems for extremophiles: one is the pHsal series for the archaeon Halobacterium salinarum,2 and the other is the Geobacillus plasmid set demonstrated on Geobacillus thermoglucosidasius.3
The genus Thermus includes a high number of bacterial thermophilic strains isolated from both marine and terrestrial natural environments and from man-made thermal facilities.4 In addition to their use as a source of thermostable enzymes of biotechnological interest, the strains of Thermus thermophilus (Tth), HB27 and HB8, have been used for a long time as model organisms of life at a high temperature, as test and selection systems for thermostable enzymes, and as sources of proteins and protein complexes for structural biology studies.5 Both Tth strains can grow from 55 to 80 °C, have a small genome consisting of a chromosome of around 2 Mb, accompanied by at least one megaplasmid (0.23 Mb), and another smaller plasmid for the HB8 strain.6 Their genome has a high GC content (69%), high coding density (95%, 2200 genes), and a low number of paralog genes. Compared to other extreme thermophiles, Tth strains grow fast and reach high cell densities in rich liquid medium under aerobic conditions (40 min doubling time at 70 °C). They can utilize a variety of saccharides and amino acids as carbon sources and form individual colonies on agar plates, thus allowing the isolation of clonal populations. In addition, the most useful characteristic of Tth as a laboratory model is the constitutive expression of a complex natural competence apparatus (NCA), which, among other components, includes type IV-like pili, which make these strains easy to transform with very high efficiency.7 Together with that, the strain HB27 shows also high proficiency in a process similar to conjugation, named transjugation, that occurs in two steps, pull and push, requiring the NCA in the recipient strain (pull) and a DNA donation system based on a DNA translocase encoded within a small operon of a mobile element (push).8
The existing toolkit for genetic manipulation of Tth is versatile and continues to expand. Plasmids, with different origins of replication,9,10 have been described, as well as three thermostable antibiotic resistances;11−13 one inducible (Pnar)14 and several constitutive promoters;15,16 a number of reporter genes, including those of a thermostable superfolder GFP,17 a β-galactosidase,14 the phytoene synthase,18 and others; and three counterselection markers.19 More recently, the Cre-lox system has also been added to the Tth toolbox.20 However, up to now, a modular system for Tth equivalent to those available for other microorganisms has not been developed.
Likewise, the accurate regulation of gene expression of both endogenous and heterologous genes in Tth, while potentially quite useful, has not been systematically addressed. A number of bifunctional promoters that work constitutively in Escherichia coli and in Tth16,21 have been described. However, in a workflow consisting of library-making in E. coli, to obtain a high number and diversity collections of genes, with subsequent transfer to Tth to screen for thermostable proteins, the best strategy would be to ensure expression in Tth while, at the same time, reducing it as much as possible in E. coli to avoid plasmid instability and/or the negative effect on representation produced by toxicity of exogenous proteins.
The work presented here describes the construction of a modular plasmid system that allows the á la carte assembly into a single plasmid of two replicative origins, up to three selection markers active in a wide temperature range, promoters, and cargo genes. This plasmid system will flexibilize the work with T. thermophilus and simultaneously allow for the bidirectional transfer to or from an appropriate mesophilic host. We also develop a series of RNA thermosensors that allow the finely regulated expression of a reporter superfolder citrine version of GFP (IFP) in Tth, while at the same time, the RNA structure keeps the expression turned off in a mesophilic host.
Results and Discussion
Design and Construction of the Modular pMoT Plasmid
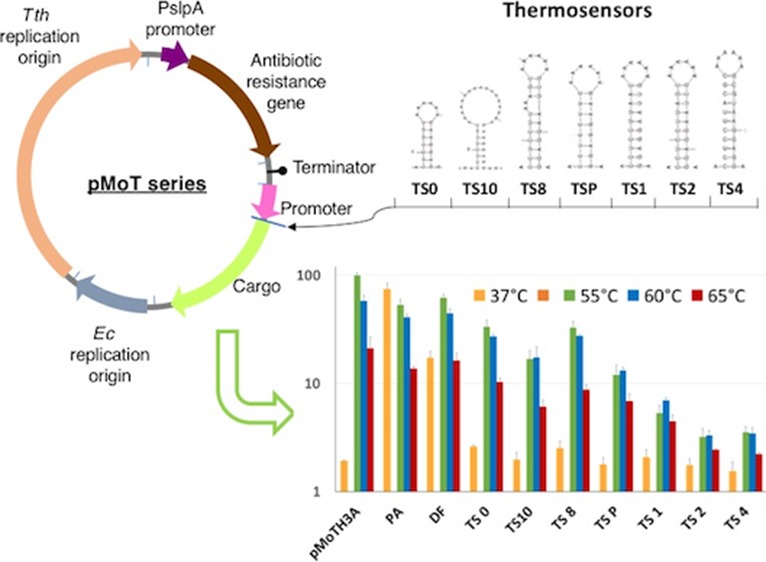
To assemble the modular plasmid chassis pMoT (plasmid Modular Thermus) (Figure 1), we started, as a first module, with the robust replication origin from pMK189,22 (pMY1 origin, pEM2S minimal origin construct, Figure 1B, see the Supporting Information for a list of plasmids and strains) that comprises the coding sequence for the replication origin type RepA binding protein and the origin itself (see Table S4 for sequences). We included two pairs of unique enzyme restriction sites flanking the replication origin: PstI and PacI at the 3′ end of Tth replication origin and AatII and NotI sites at the 5′ end. As an alternative origin of replication for Thermus spp., we have set up a 2400-bp fragment containing the origin of replication from plasmid pTT8.10 A second module for antibiotic selection follows the PacI site and includes the strong PslpA promoter, active both in Tth and E. coli,23 that drives the expression of the antibiotic resistance gene of choice, followed in all cases by an AscI site. Three thermostable antibiotic resistance cassettes are available that confer resistance to kanamycin, hygromycin B (a more thermostable version, Bosch et al.99), and bleomycin B (Figure 1B).11−13 The three markers work robustly and stably in both Tth at 65 °C and E. coli at 37 °C, which makes the addition of a dedicated antibiotic resistance for the mesophilic host unnecessary, keeping the plasmid at a minimum size. A strong Rho-independent transcriptional terminator from the SlpA gene is located after the antibiotic resistance cassette, and the cargo module follows this terminator. We have tested several promoters (see below) and selected the nqo promoter (Pnqo)24 as the standard for cargo expression because it is very active in Tth but essentially inactive in E. coli (see below). The strength of Pnqo is comparable to that of the promoter PslpA or that of the 16S rRNA, both commonly considered to be very active in bacteria25,26 (see below). An EcoRI site follows Pnqo and then a ribosome binding site and an NdeI site that can be used for the expression of any cloned gene from its ATG codon. The cargo gene can be cloned between EcoRI (with its own RBS) or NdeI and HindIII sites. As a reporter, we have used a superfolder citrine fluorescent protein (sIFP) (see the Supporting Information), but as mentioned above, we also have available genes for β-galactosidase, alkaline phosphatase, and β-carotene synthesis and a thermostabilized version of esterase I from Pseudomonas fluorescens (Maté et al.100) as reporters (Figure 1B). Finally, a module for replication in mesophilic hosts is included between the HindIII and NotI sites. In addition to the pUC-derived replicon with a high copy number in E. coli, the pBBR Gram-negative broad host range replicon from the pSEVA131 vector has been also tested.27 We have also constructed a series of plasmids with a polylinker (linker A) in place of the promoter-cargo modules (Figure 1C), and we have constructed a series of plasmids with Pnqo in front of polylinkers, with RBSs directing expression in the three reading frames for library cloning and testing (linkers 1, 2, and 3, Figure 1C).
Figure 1.
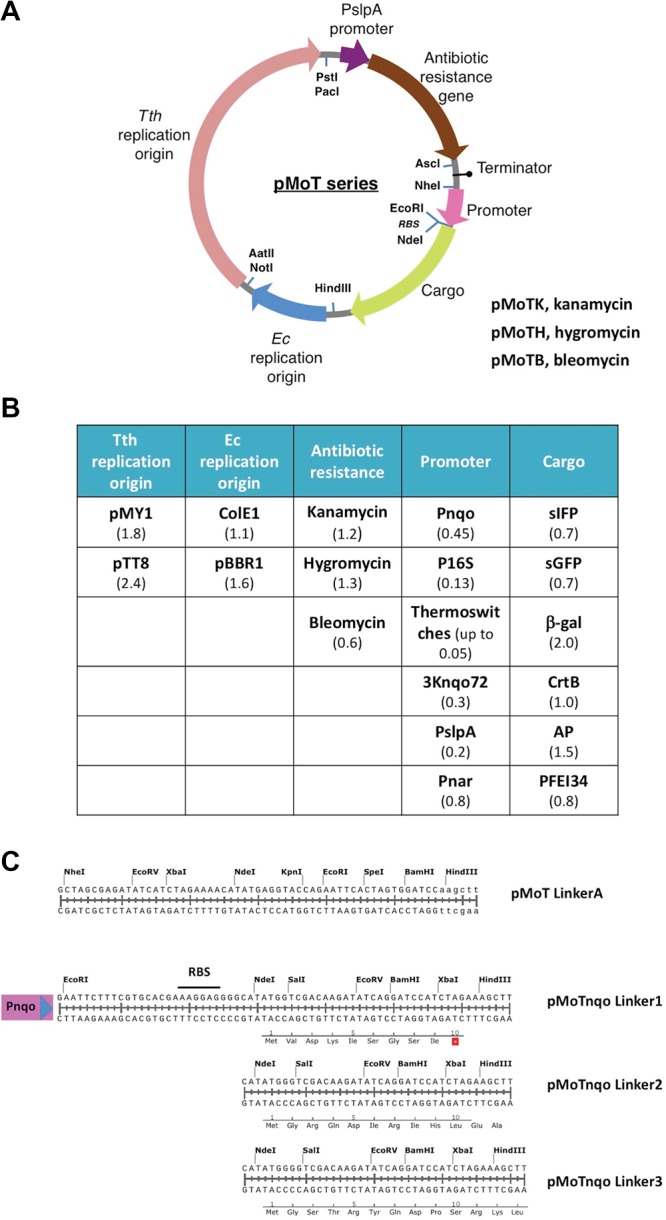
(A) Diagram of the pMoT series plasmid system. PslpA is the 177-bp Tth SlpA gene promoter. Indicated with arrows of different colors are the component modules; also shown are the restriction sites connecting the modules. Tth, T. thermophilus; Ec, E. coli. See Table S4 for a list of the module parts. (B) Summary of pMoT component parts. Pnqo, promoter for the nqo gene; P16S, promoter for the Tth 16S RNA; thermosensors, translation control structures described in this work; 3Knqo72, fusion of promoter nqo72 plus promoter 3K; PslpA, promoter for S-layer A gene; Pnar, nitrate reductase promoter; sIFP, superfolder citrine GFP; sGFP, superfolder GFP; β-gal, β-galactosidase; CrtB, β-carotene synthesis gene; AP, alkaline phosphatase; PFEI, esterase I variety 34 from P. fluorescens. See the Supporting Information for references. Length of the DNA fragments, in Kbs, is indicated between parentheses. (C) Sequence of the different multiple cloning sites indicating the unique restriction sites. Pnqo, promoter of the gene nqo; RBS, ribosome binding site. Linkers 1, 2, and 3 are designed to clone fragments and translate them in the three different frames; translation in the corresponding frame is shown.
Assessment of Promoters for the Expression of the Cargo Genes in T. thermophilus and E. coli
The modular plasmid was used in combination with sIFP to test a number of promoters to define their behavior in Tth (Figure 2A) and in E. coli (Figure 2B). As shown in Figure 2A, Pnqo is a strong promoter in Tth (NQO) but has no detectable activity in E. coli (Figure 2B). By contrast, a version of the Plac promoter with an incomplete lac operator (LAC) is almost inactive in Tth but is strongly expressed in E. coli. Fluorescence from similar plasmids without sIFP (pMoTK) or with sIFP without a promoter (Δprom) was almost negligible. The promoters from the 16S rRNA28 from Tth are strong in both Tth and E. coli (Figure 2A,B, bar 16S). We also tested a “three kingdom” (3K) promoter from the archaeon Haloferax volcanii, described as also active in transcription in E. coli and Saccharomyces cerevisiae(29) with the idea of having a multikingdom promoter operating in our system. However, the 3K promoter showed low levels of transcription in E. coli and extremely low levels in Tth (Figure 2A,B, bar 3K) (see below).
Figure 2.
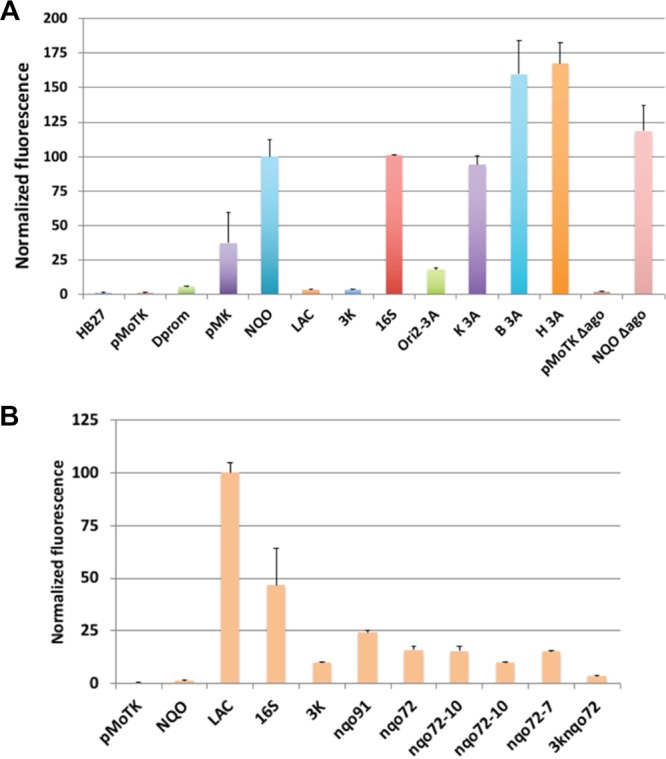
Characterization of different promoters in the pMoT system. (A) Fluorescence levels measured for the indicated promoters transformed into Tth. HB27, Tth HB27 strain with no plasmid; pMoTK, plasmid without promoter and sIFP gene; Δprom, construct without promoter; pMK, pMKnqosGFP; NQO, pMoTK with Pnqo driving sIFP; LAC, pMoTK with Lac promoter driving sIFP; 3K, idem with 3K promoter; 16S, idem with rRNA 16S promoter; Ori2-3A, pMoTK with nqo 3A promoter and pTT8 origin of replication; K 3A, pMoTK with nqo 3A promoter; B 3A, pMoTB with nqo 3A promoter; H 3A, pMoTH with nqo 3A promoter; C + Δago, pMoTK with Pnqo-sIFP transformed in Δago strain; C-Δago, pMoTK transformed in Δago strain. (B) Fluorescence levels measured for the indicated promoters in E. coli cultured at 37 °C. nqo variants as indicated in Figure 3B.
The combination of the presence of an antibiotic and the expression of antibiotic resistance could have an effect on the expression levels of the cargo gene. Therefore, we tested fluorescence generated from plasmids pMoTK, pMoTB, and pMoTH, endowing resistance to kanamycin, hygromycin, and bleomycin, respectively. For these assays, the sIFP gene was expressed from Pnqo3A, a smaller derivative of Pnqo with essentially the same activity. As shown in Figure 2A, pMoTB and pMoTH produce in Tth about 50% more fluorescence than pMoTK (bars K 3A, B 3A, and H 3A). On the other hand, the expression from Pnqo3A within a pMoTK derivative with pTT8 replication origin is reduced to 17% compared with the same construct with the pMY1 origin (bars Ori 2-3A vs K 3A), in accordance with the fact that the pTT8 origin yields lower copy number vectors.10 Finally, the putative effects of the absence of the Argonaute protein in the expression was assayed as Δago mutants are commonly used for cloning purposes in Tth due to their higher transformation efficiencies.30 As shown by the corresponding bar (Figure 2A, bar 3A ΔAgo), the expression levels in this mutant is similar to those in the corresponding wild-type strain.
Analysis of the Pnqo Elements in T. thermophilus and E. coli
The data above show that the Pnqo promoter is only functional in Tth, showing no activity at all in E. coli (Figure 2A,B), and so, it matches the profile for a promoter that ensures good expression in Tth while preventing unwanted activity in E. coli. To further study Pnqo, we performed serial deletions from its 5′ end, and their promoter activities were assayed in Tth and E. coli. As shown in Figure 3A, deletion of regions upstream of position −72 from the ATG start codon produced nonsignificant variations in the activity respect to the original Pnqo, which extended to position −446. Interestingly, deletions nqo91 and nqo72 increased the expression by around 10-fold in E. coli with respect to the intact promoter (Figure 3A), probably due to the new upstream regions added in the construction (Figure 3B). Further deletions down to position −57 or −33 produced the complete loss of promoter activity in Tth, supporting the presence of relevant sequences for the binding of the Tth RNA polymerase between positions −72 and −57. Actually, the sequence between these positions is similar to an extended −10 promoter box consensus (Figure 3B). To verify if this was the case, we made point mutations on that putative box and also mutated a putative −35 box (TTGCGC) found between positions −82 and −76, all in the context of the nqo91 deletion mutant that retained almost full activity. As it is shown in Figure 3A, mutations leading to a nonconsensus −35 box (column nqo91-35) did not produce a decrease in the promoter activity, whereas any of the pairs of mutations on the extended −10 box (nqo91E-10, nqo91-10, nqo91-7) strongly reduced the promoter activity. Since the nqo72 version of the promoter is also active in E. coli, we combined the nqo72 version with the extended −10 region mutants, and we observed that, while the activity is still strongly reduced in Tth (nqo72-E10 to nqo72-7), it is still active, although decreased by 50%, in E. coli (Figure 2B). These results suggest that the Pnqo promoter in Tth is heavily reliant on an extended −10 TATA element, while on the other hand, for expression in E. coli, a −35 box would be needed, whereas the promoter strength in E. coli is not so much dependent on the extended −10 region.
Figure 3.
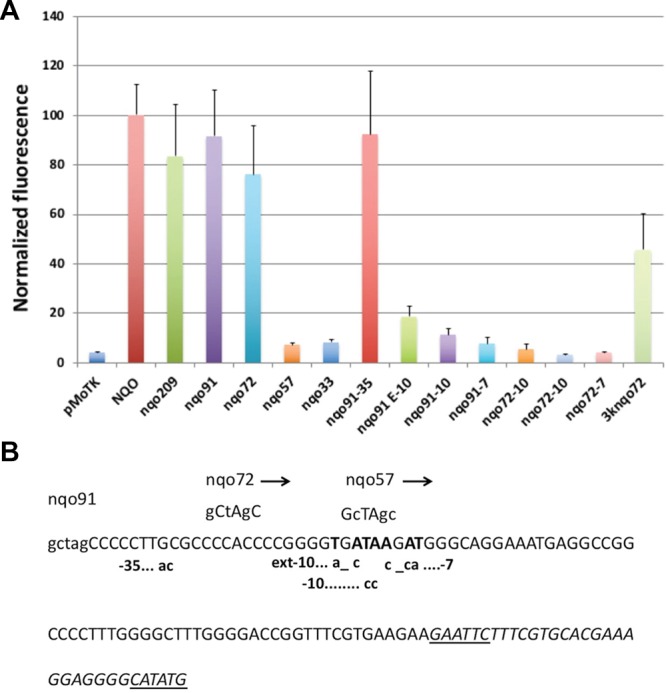
Characterization of different promoters in the pMoT system. (A) Fluorescence levels measured for the indicated promoters in Tth cultured at 65 °C. nqo209, nqo91, nqo72, nqo57, and nqo33 denote the remaining sequence of the nqo promoter at the 3′ end. nqo91-35, mutations in the proposed −35 box constructed on Pnqo91; nqo91E-10, mutations in the extended −10 region; nqo91-10, mutations in the −10 region; Nqo91-7 mutations in the −7 region; nqo72E-10, nqo72-10, nqo72-7, same mutations introduced on Pnqo72; 3Knqo72, hybrid 3K-nqo72 promoter. (B) Sequences of nqo promoter mutants.
Finally, since we had delimitated a minimal promoter with good activity in Tth and significant activity in E. coli, we replaced the region described as needed for expression in E. coli in the 3K promoter by the nqo72 derivative while keeping the sequences required for expression in H. volcanii and S. cerevisiae. The new 3Knqo72 promoter showed 40% of the activity relative to wild-type Pnqo (Figure 3A) in Tth but still had low activity in E. coli. This suggests that the 3Knqo72 promoter could be a promising tool for a bifunctional Thermus-Haloferax promoter.
The Use of Thermosensors for High-Temperature Protein Expression
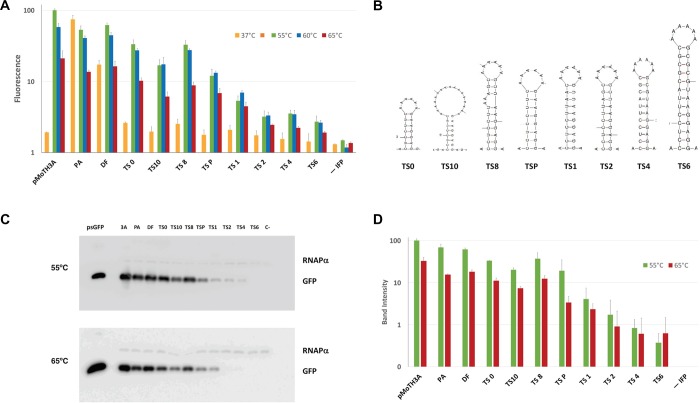
Given the different growth temperatures of E. coli and Tth, we reasoned that RNA thermosensors could serve as adequate controllers of gene expression in the context of the pMoT plasmids in two ways: first, by granting expression in Tth while repressing it in E. coli and, second, by allowing the fine-tuning of expression in Tth just by changing the growth temperature to induce the opening or closing of the relevant thermosensor. Inspired on a previous report,31 we designed a series of RNA thermosensors acting by RBS occlusion in stem-loop structures with melting temperatures, derived from ΔG, ranging from 50 to 70 °C (see Figure 4B and Figure S1 for predicted structures and ΔG). The thermosensors were inserted between hygromycin B and the sIFP gene in bicistronic constructions expressed from the PslpA promoter in a pMoTH. In these constructs, expression of hygromycin phosphotransferase should be unaffected, while the sIFP gene should be regulated by the thermosensors. The thermosensor-bearing plasmids were transformed into the Δago mutant strain. The fluorescence measures obtained under different conditions are shown in Figure 4A, and confirmatory immunodetection of sIFP is shown in Figure 4C, with quantization of three representative experiments performed at 55 and 65 °C, as presented in Figure 4D. A linear correlation between fluorescence and Western blot data is shown in Figure S2. It can be observed that expression in E. coli is almost completely abolished by the presence of any of the thermosensors (orange columns), whereas the control with polyA at the same place (PA) or without a thermosensor (direct fusion (DF)) showed strong or intermediate fluorescence, respectively. This indicates that the thermosensors can be used to restrict expression out from a mesophilic host while having good expression in Tth. On the other hand, the thermosensors also produced repression of expression in Tth with respect to the controls, following approximately the pattern expected due to the ΔG’s associated to the different structures. However, unexpectedly, for a given thermosensor, there is an apparent increase in expression at 55 and 60 °C with respect to the expression at 65 °C. Also, this effect is also observed in the control plasmid pMoTH 3A when IFP is driven by the Pnqo promoter without any thermosensor. This effect is contrary to the behavior predicted for the different thermosensors that should tend to derepress expression with the increase in temperature. At present, we do not know the reasons for this increased expression at 55 and 60 °C. In any event, the differences in expression levels obtained with the thermosensor series span about 2 orders of magnitude, and the range of thermosensors can be used to regulate expression from very low levels (thermosensors 2, 4, and 6) through medium levels (P and 1) to high levels (0, 10, and 8). The results obtained with this thermosensor series are similar or, in some cases, better than those in reported studies, yielding induction folds of 10 to 35 times with respect to the uninduced state.32,33 We get a maximum of 13-fold induction of expression of IFP when comparing normalized expression in E. coli with normalized expression in Tth (TS8) and, by Western blot quantization, some 180-fold differences between the construct and the highest expression (PA) and the lowest one (TS6). While, additionally, in our study, we use the thermosensors to repress expression in a mesophilic host and to regulate protein output with the same vector in a thermophilic model, this had not been reported before. On the other hand, our system would be susceptible of improvement since better systems of induction of expression for Tth are desirable, and further work based on this set of thermosensors could lead to easy-to-use expression systems.
Figure 4.
Characterization of thermosensors in the pMoT system. (A) Fluorescence levels measured for the indicated constructions with thermosensors (0, 10, 8, P, 1, 2, 4, 6), constructs without thermosensors (polyA (PA) and direct fusion (DF)), control lacking sIFP gene (-IFP), and Pnqo 3A driving sIFP expression (3A). Orange bars, expression in E. coli at 37 °C; green bars, expression in Tth at 55 °C; blue bars, expression in Tth at 60 °C; red bars, expression in Tth at 65 °C. (B) The predicted RNA structure is shown for each thermosensor (see the Supporting Information). (C) Western blot detection of sIFP from the corresponding constructs from Tth cultures maintained at 65 or 55 °C. psGFP, lane loaded with 50 ng of purified sGFP. (D) Quantization of Western blot sIFP bands. Labels are the same as in (A).
The thermosensors presented here, as they are based on physical properties, could also be utilized to regulate gene expression not only in Tth but also in other thermophilic organisms. We expect that the thermosensors, together with the modular plasmid system described here, will help build and standardize future work and developments on Tth biology.
Materials and Methods
Bacterial Strains and Growth Conditions
Bacterial strains and plasmids used in this work together with growth conditions are listed in the Supporting Methods (Supporting Information).
Nucleic Acid Manipulation
Primers and plasmids used in this work are listed in Tables S2 and S3, respectively. Plasmid construction, purification, restriction analysis, and DNA sequencing were performed by standard methods. DNA was amplified by PCR using 1 U/100 μL of DNA polymerase PfuUltra II (Roche Molecular Biochemicals) in its recommended buffer with 200 μM dNTP mixture and 1 μM of each primer (Sigma-Aldrich).
Plasmid Construction
The parts used to assemble pMoT plasmids and their sequences are listed in Table S4. Parts were PCR-amplified using the primers listed in Table S2, adding the appropriate restriction sites, and plasmids were assembled by ligation to produce the plasmids listed in Table S3. Construction was started with pMoTK110. All the constructs were confirmed by sequencing. Additional details about constructs can be found in the Supporting Methods.
Promoter Activity Assays
The promoter regions to be tested were cloned between sites NheI and EcoRI in the plasmid pMoTK110, driving the expression of superfolder sIFP. Expression from these promoters was assayed on transformed T. thermophilus strains after growth of overnight 10 mL cultures to saturation at 65 °C followed by dilution to OD600 = 0.05 and then culture for 24 h at the indicated temperatures. Fluorescence measures were performed on 1.66 × 108 cells, washed twice in PBS, and resuspended in 200 μL using a FLUOstar Optima (BMG Labtech, Germany) fluorimeter. Measures were performed from three biological replicates and at least three technical replicates for each condition.
Western Blotting
Tth cells transformed with the corresponding plasmids were cultured as described above, and then total cell extracts were prepared by resuspending 109 cells in 200 μL of loading buffer and boiling for 15 min. The extracts were analyzed by SDS-PAGE, electrotransferred to a PVDF membrane, and detected with specific rabbit antisera (anti-GFP polyclonal (Chromotek) and anti-Bacillus subtilis RNA polymerase α subunit as a control (the later, a kind gift from Margarita Salas). The membranes were developed using horseradish peroxidase-labeled anti-rabbit antibodies and bioluminescence assay (ECL, Amersham International). Western blot band quantization was performed with an Amersham Imager 680 chemiluminescence reader using the ImageJ image analysis software.34
Acknowledgments
This work was supported by the European Union grant H2020-BIOTEC-2014-2015/H2020-LEIT-BIO-2015-1 and grant BIO2016-77031R from the Spanish Ministry of Science, Innovation and Universities (MICIU). An institutional grant to CBMSO from Fundación Ramón Areces and an FPI contract to C.V. from the MCIU are also acknowledged.
Glossary
Abbreviations
- pMoT
plasmid modular for Thermus
- Tth
Thermus thermophilus
- E. coli
Escherichia coli
- bp
base pair
- GFP
green fluorescent protein
- sIFP
citrine fluorescent protein
- DF
direct fusion
- PA
polyA
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsomega.9b02107.
Table S1, strain list; Table S2, plasmid list; Table S3, oligonucleotide list; Table S4, list of component sequences; Table S5, references for component parts; Supporting Methods; nucleotide sequence and map of plasmid pMoTH150; Figure S1; and Figure S2 (PDF)
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Nora L. C.; Westmann C. A.; Martins-Santana L.; Alves L. F.; Monteiro L. M. O.; Guazzaroni M.-E.; Silva-Rocha R. The art of vector engineering: towards the construction of next-generation genetic tools. Microb. Biotechnol. 2019, 12, 125–147. 10.1111/1751-7915.13318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva-Rocha R.; Pontelli M. C.; Furtado G. P.; Zaramela L. S.; Koide T. Development of New Modular Genetic Tools for Engineering the Halophilic Archaeon Halobacterium salinarum. PLoS One 2015, 10, e0129215 10.1371/journal.pone.0129215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeve B.; Martinez-Klimova E.; de Jonghe J.; Leak D. J.; Ellis T. The Geobacillus Plasmid Set: A Modular Toolkit for Thermophile Engineering. ACS Synth. Biol. 2016, 5, 1342–1347. 10.1021/acssynbio.5b00298. [DOI] [PubMed] [Google Scholar]
- da Costa M. S.; Rainey F.; Nobre M. F.. The Genus Thermus and Relatives. In The Prokaryotes; 3rd ed.; Dworkin M., Ed. Springer: 2006, Vol. 7, pp 797–812. [Google Scholar]
- Cava F.; Hidalgo A.; Berenguer J. Thermus thermophilus as biological model. Extremophiles 2009, 13, 213–31. 10.1007/s00792-009-0226-6. [DOI] [PubMed] [Google Scholar]
- Henne A.; Brüggemann H.; Raasch C.; Wiezer A.; Hartsch T.; Liesegang H.; Johann A.; Lienard T.; Gohl O.; Martinez-Arias R.; Jacobi C.; Starkuviene V.; Schlenczeck S.; Dencker S.; Huber R.; Klenk H.-P.; Kramer W.; Merkl R.; Gottschalk G.; Fritz H.-J. The genome sequence of the extreme thermophile Thermus thermophilus. Nat. Biotechnol. 2004, 22, 547–53. 10.1038/nbt956. [DOI] [PubMed] [Google Scholar]
- Friedrich A.; Hartsch T.; Averhoff B. Natural transformation in mesophilic and thermophilic bacteria: identification and characterization of novel, closely related competence genes in Acinetobacter sp. strain BD413 and Thermus thermophilus HB27. Appl. Environ. Microbiol. 2001, 67, 3140–8. 10.1128/AEM.67.7.3140-3148.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blesa A.; Baquedano I.; Quintáns N. G.; Mata C. P.; Castón J. R.; Berenguer J. The transjugation machinery of Thermus thermophilus: Identification of TdtA, an ATPase involved in DNA donation. PLoS Genet. 2017, 13, e1006669 10.1371/journal.pgen.1006669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Grado M.; Castán P.; Berenguer J. A high-transformation-efficiency cloning vector for Thermus thermophilus. Plasmid 1999, 42, 241–5. 10.1006/plas.1999.1427. [DOI] [PubMed] [Google Scholar]
- Takayama G.; Kosuge T.; Maseda H.; Nakamura A.; Hoshino T. Nucleotide sequence of the cryptic plasmid pTT8 from Thermus thermophilus HB8 and isolation and characterization of its high-copy-number mutant. Plasmid 2004, 51, 227–37. 10.1016/j.plasmid.2004.01.003. [DOI] [PubMed] [Google Scholar]
- Hoseki J.; Yano T.; Koyama Y.; Kuramitsu S.; Kagamiyama H. Directed evolution of thermostable kanamycin-resistance gene: a convenient selection marker for Thermus thermophilus. J. Biochem. 1999, 126, 951–6. 10.1093/oxfordjournals.jbchem.a022539. [DOI] [PubMed] [Google Scholar]
- Nakamura A.; Takakura Y.; Kobayashi H.; Hoshino T. In vivo directed evolution for thermostabilization of Escherichia coli hygromycin B phosphotransferase and the use of the gene as a selection marker in the host-vector system of Thermus thermophilus. J. Biosci. Bioeng. 2005, 100, 158–63. 10.1263/jbb.100.158. [DOI] [PubMed] [Google Scholar]
- Brouns S. J. J.; Wu H.; Akerboom J.; Turnbull A. P.; de Vos W. M.; van der Oost J. Engineering a selectable marker for hyperthermophiles. J. Biol. Chem. 2005, 280, 11422–11431. 10.1074/jbc.M413623200. [DOI] [PubMed] [Google Scholar]
- Moreno R.; Zafra O.; Cava F.; Berenguer J. Development of a gene expression vector for Thermus thermophilus based on the promoter of the respiratory nitrate reductase. Plasmid 2003, 49, 2–8. 10.1016/S0147-619X(02)00146-4. [DOI] [PubMed] [Google Scholar]
- Maseda H.; Hoshino T. Screening and analysis of DNA fragments that show promoter activities inThermus thermophilus. FEMS Microbiol. Lett. 1995, 128, 127–34. 10.1111/j.1574-6968.1995.tb07511.x. [DOI] [PubMed] [Google Scholar]
- Park H. S.; Kilbane J. J. II Gene expression studies of Thermus thermophilus promoters PdnaK, Parg and Pscs-mdh. Lett. Appl. Microbiol. 2004, 38, 415–22. 10.1111/j.1472-765X.2004.01512.x. [DOI] [PubMed] [Google Scholar]
- Cava F.; de Pedro M. A.; Blas-Galindo E.; Waldo G. S.; Westblade L. F.; Berenguer J. Expression and use of superfolder green fluorescent protein at high temperatures in vivo: a tool to study extreme thermophile biology. Environ. Microbiol. 2008, 10, 605–13. 10.1111/j.1462-2920.2007.01482.x. [DOI] [PubMed] [Google Scholar]
- Fujita A.; Misumi Y.; Honda S.; Sato T.; Koyama Y. Construction of new cloning vectors that employ the phytoene synthase encoding gene for color screening of cloned DNA inserts in Thermus thermophilus. Gene 2013, 527, 655–62. 10.1016/j.gene.2013.06.069. [DOI] [PubMed] [Google Scholar]
- Carr J. F.; Danziger M. E.; Huang A. L.; Dahlberg A. E.; Gregory S. T. Engineering the genome of Thermus thermophilus using a counterselectable marker. J. Bacteriol. 2015, 197, 1135–44. 10.1128/JB.02384-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Togawa Y.; Nunoshiba T.; Hiratsu K. Cre/lox-based multiple markerless gene disruption in the genome of the extreme thermophile Thermus thermophilus. Mol. Genet. Genomics 2018, 293, 277–291. 10.1007/s00438-017-1361-x. [DOI] [PubMed] [Google Scholar]
- Fujiwara K.; Sawamura T.; Niwa T.; Deyama T.; Nomura S. M.; Taguchi H.; Doi N. In vitro transcription-translation using bacterial genome as a template to reconstitute intracellular profile. Nucleic Acids Res. 2017, 45, 11449–11458. 10.1093/nar/gkx776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Grado M.; Lasa I.; Berenguer J. Characterization of a plasmid replicative origin from an extreme thermophile. FEMS Microbiol. Lett. 1998, 165, 51–7. 10.1111/j.1574-6968.1998.tb13126.x. [DOI] [PubMed] [Google Scholar]
- Faraldo M. L.; de Pedro M. A.; Berenguer J. Cloning and expression in Escherichia coli of the structural gene coding for the monomeric protein of the S layer of Thermus thermophilus HB8. J. Bacteriol. 1991, 173, 5346–51. 10.1128/jb.173.17.5346-5351.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch S.; Quesada J.; Sanchez E.; Lim J.; Berenguer J.; Hidalgo A.. Engineered hygromycin phosphotransferase for selection in extreme thermophiles at temperatures above 70 °C. To be submitted for publication. [Google Scholar]
- Cava F.; Laptenko O.; Borukhov S.; Chahlafi Z.; Blas-Galindo E.; Gómez-Puertas P.; Berenguer J. Control of the respiratory metabolism of Thermus thermophilus by the nitrate respiration conjugative element NCE. Mol. Microbiol. 2007, 64, 630–46. 10.1111/j.1365-2958.2007.05687.x. [DOI] [PubMed] [Google Scholar]
- Faraldo M. M.; de Pedro M. A.; Berenguer J. Sequence of the S-layer gene of Thermus thermophilus HB8 and functionality of its promoter in Escherichia coli. J. Bacteriol. 1992, 174, 7458–62. 10.1128/jb.174.22.7458-7462.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul B. J.; Ross W.; Gaal T.; Gourse R. L. rRNA transcription inEscherichia coli. Annu. Rev. Genet. 2004, 38, 749–70. 10.1146/annurev.genet.38.072902.091347. [DOI] [PubMed] [Google Scholar]
- Mate D.; Rivera N.; Sanchez-Freire E.; Ayala J.; Berenguer J.; Hidalgo A.. Thermostability enhancement of the Pseudomonas fluorescens esterase I by in vivo folding selection in Thermus thermophilus. Biotechnol. Bioeng. Submitted for publication. [DOI] [PubMed] [Google Scholar]
- Silva-Rocha R.; Martínez-García E.; Calles B.; Chavarría M.; Arce-Rodríguez A.; de Las Heras A.; Páez-Espino A. D.; Durante-Rodríguez G.; Kim J.; Nikel P. I.; Platero R.; de Lorenzo V. The Standard European Vector Architecture (SEVA): a coherent platform for the analysis and deployment of complex prokaryotic phenotypes. Nucleic Acids Res. 2013, 41, D666–D675. 10.1093/nar/gks1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann R. K.; Erdmann V. A. Thermus thermophilus 16S rRNA is transcribed from an isolated transcription unit. J. Bacteriol. 1989, 171, 2933–2941. 10.1128/jb.171.6.2933-2941.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.; Huang Y. P.; Shen P. The 492-bp RM07 DNA fragment from the halophilic Archaea confers promoter activity in all three domains of life. Curr. Microbiol. 2003, 47, 388–394. 10.1007/s00284-003-4034-4. [DOI] [PubMed] [Google Scholar]
- Swarts D. C.; Jore M. M.; Westra E. R.; Zhu Y.; Janssen J. H.; Snijders A. P.; Wang Y.; Patel D. J.; Berenguer J.; Brouns S. J. J.; van der Oost J. DNA-guided DNA interference by a prokaryotic Argonaute. Nature 2014, 507, 258–261. 10.1038/nature12971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neupert J.; Karcher D.; Bock R. Design of simple synthetic RNA thermometers for temperature-controlled gene expression in Escherichia coli. Nucleic Acids Res. 2008, 36, e124. 10.1093/nar/gkn545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury S.; Ragaz C.; Kreuger E.; Narberhaus F. Temperature-controlled structural alterations of an RNA thermometer. J. Biol. Chem. 2003, 278, 47915–21. 10.1074/jbc.M306874200. [DOI] [PubMed] [Google Scholar]
- Sadler F. W.; Dodevski I.; Sarkar C. A. RNA Thermometers for the PURExpress System. ACS Synth. Biol. 2017, 7, 292–296. 10.1021/acssynbio.7b00294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C. A.; Rasband W. S.; Eliceiri K. W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–5. 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.