Figure 2.
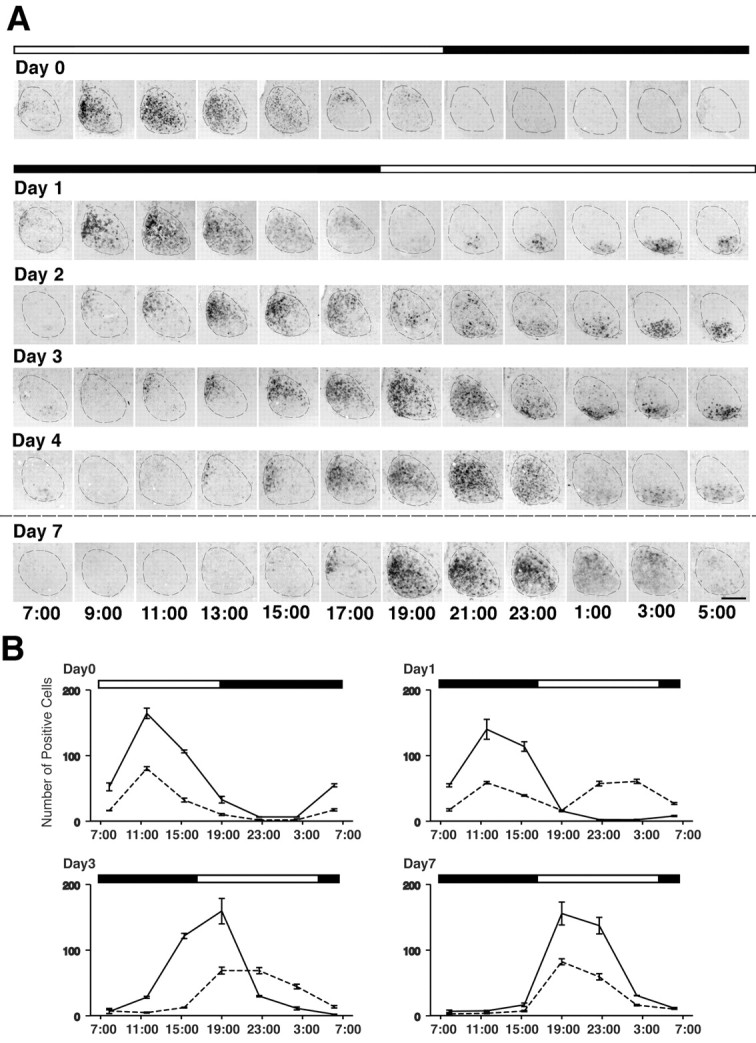
Detailed time course of rPer1 expression in the SCN after a delaying shift in the light/dark cycle. A, Representative coronal brain sections of the SCN processed for rPer1 in situ hybridization. The top black-and-white bar indicates the original LD cycle, and the bottom bar shows the new light cycle. The dashed line indicates the SCN. Clock time is displayed below each column of photomicrographs. On the day before the photoperiod shift (Day 0), the VLSCN and DMSCN are synchronized with respect to their rPer1 expression. On the days after the shift, the two subregions are initially desynchronized but eventually become completely resynchronized by Day 7. Labeled cells in the VLSCN are reset to the new photoperiod by Day 1, whereas this process takes several days for the cells in the DMSCN. Scale bar, 200 μm. B, Graphs showing the number of SCN cells expressing rPer1 mRNA in the DMSCN (solid line) or the VLSCN (dashed line) under entrained conditions (Day 0) or in the days after a 10 hr photoperiod shift (Day 1, Day 3, Day 7). Each time point indicates the mean ± SEM of three rats.
