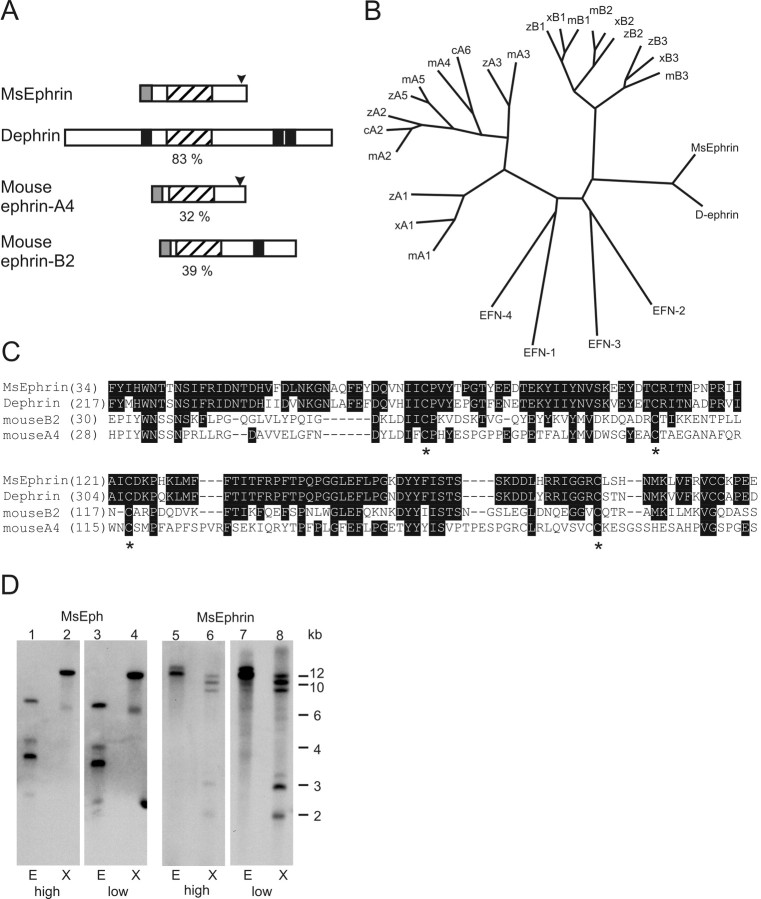
Figure 2.
Alignment of MsEphrin with other members of the ephrin family and genomic Southern blot analyses for MsEph and MsEphrin. A, Structural comparison of MsEphrin with Dephrin and a vertebrate A class and B class ephrin. Numbers in percentages indicate the amino acid identity of the ephrin core domain of MsEphrin to those in other ephrins. Conserved ephrin core domains (hatched boxes), GPI-anchoring motifs (arrowheads), transmembrane segments (black boxes), and secretion signal peptides (shaded boxes) are indicated. B, Phylogenetic tree analysis of the ephrin core domains. Species are indicated as c, chick; m, mouse; x, Xenopus laevis; z, zebrafish. C. elegans ephrins, EFN1-4, are all GPI-anchored. See also the legend for Figure 1, C and D. C, Amino acid alignment of the ephrin core domain. Identical residues are printed in white lettering on black background. Asterisks indicate the conserved cysteine residues that have been crystallographically shown to form disulfide bonds (Himanen et al., 2001; Toth et al., 2001). Numbers are amino acid positions. Sequences of Dephrin, mouse ephrin-A4, and -B2 were obtained from GenBank accession numbers AF216287, U90663, and U16819, respectively. D, Genomic Southern blot analyses for MsEph and MsEphrin. Lanes 1-4 are for MsEph and lanes 5-8 are for MsEphrin. Restriction endonucleases used are indicated as E, EcoRI and X, XbaI. Hybridization-washing conditions are indicated as either high-stringency (high) or low-stringency (low) at the bottom. Note that two bands clearly seen at 2 and 3 kb in MsEphrin low-stringency condition (lane 8) were also detected, albeit faintly, in the high-stringency condition (lane 6).