Abstract
Toll-like receptors (TLRs) are key molecules in innate immunity that recognize a variety of pathogen-associated molecular patterns. Activation of TLRs by their agonists initiates several signaling cascades, which eventually result in the expression of immune modifiers. Despite the fact that MCPIP1 is reported as an important immune regulator involved in macrophage activation, modulation of its expression by all known TLR agonists has never been documented. In this study, we present for the first time that in human monocyte-derived macrophages all TLR agonists, except CpG, markedly induced the expression of MCPIP1. The level of the induced transcript, as well as the protein and time of their appearance varied depending on the agonist. Furthermore, we confirmed the strong and differential upregulation of MCPIP1 during bacteria, virus and fungus infection. MCPIP1 belongs to a group of early-response genes; however, in the present study, we show for the first time the sustained high level of MCPIP1 expression during long-term Staphylococcus aureus infection. Taken together, our results implicate MCPIP1 as a potent regulator of innate immunity, which can be strongly engaged in the pathogenesis of acute and chronic infective diseases.
Key Words: MCPIP1, Toll-like receptors, Innate immunity, Infection
Introduction
Toll-like receptors (TLRs) are a group of pattern recognition receptors that play an essential role in host defense against a wide range of pathogenic microorganisms [1]. TLRs are thought to function by detecting the presence of distinct components unique to microorganisms and subsequently activate different gene programs, which promote various innate and adaptive immune responses. Thus far, ten TLRs have been identified in humans; however, their distribution is varied. TLRs are members of the interleukin (IL)-1R superfamily and share a similar signaling cascade. The cascade involves activation of the adapter molecule MyD88, which forms an initial signaling complex with IRAK4 and TRAF6. Formation of this complex results in activation of a signaling cascade that leads to activation of the MAP kinases, such as ERK, JNK and p38, as well as transcription factors such as nuclear factor (NF)-ĸB and activator protein 1. Despite shared signaling pathways, engagement of different TLRs elicits a spectrum of immune responses because TLRs differ in their cellular localization and ligand specificity, as well as in the rate, intensity and/or efficiency of activation of target genes, some of which involve as yet unidentified mechanisms. Therefore, the investigation of TLR-inducible genes is important for clarifying the control mechanisms of innate immune reactions.
The MCPIP1 gene (also called ZC3H12A), originally described as a gene activated by monocyte chemoattractant protein 1 (MCP-1), belongs to a group known as CCCH zinc finger genes that encompasses 55 members described in humans [2]. MCPIP1 contains a putative nuclear localization signal sequence, a single zinc finger domain with three cysteines, and one histidine for RNA binding; therefore, this protein was described originally as a transcription factor that induces expression of apoptotic gene families [2]. Further studies showed, that MCPIP1 has a PilT N terminus domain with RNase properties and regulates the stability not only of its own transcript but also of IL-6, IL-1 and IL-12p40 mRNA [3, 4]. Furthermore, recent data revealed that MCPIP1 can act as a deubiquitinase for tumor necrosis factor receptor (TNFR)-associated factors, thus negatively regulating JNK- and NF-ĸB-dependent signaling [5, 6, 7]. The newest data identify MCPIP1 as a broad suppressor of miRNA activity and biogenesis, which counteracts Dicer, a central ribonuclease in the miRNA processing system [8]. Such features predispose MCPIP1 to play an important role in both physiological and pathological processes related to inflammation. MCPIP1 gene transcription is rapidly and potently induced upon stimulation with proinflammatory molecules such as TNF-α, MCP-1, IL-1β and lipopolysaccharide (LPS) [2, 9]. This process is regulated via NF-ĸB and MAP kinase pathways [5, 7, 10]. Commitment of such crucial components of the inflammatory cell-signaling network suggests, that MCPIP1 can be upregulated by different ligand-receptor pairs, including TLRs. However, this remains to be fully elucidated, since MCPIP1 itself is a factor which critically regulates inflammation [3].
In this study, we report the first comparative analysis of the differential effects of TLR agonists on MCPIP1 mRNA and protein expression in human primary macrophages, which are reported to express all TLRs [11]. We used either natural or synthetic TLR agonists and also examined the role of viable pathogens (Staphylococcus aureus, Escherichia coli, Candida albicans, adenovirus type 1) on short- and long-term MCPIP1 regulation in infected macrophages.
Materials and Methods
Reagents
Gentamycin, SYBR Green JumpStart Taq Ready Mix, tryptic soy agar, tryptic soy broth, endotoxin (LPS), lipoteichoic acid (LTA) and poly(I:C) were from Sigma. Fetal calf serum, RPMI-1640, calcium- and magnesium-free PBS (without Ca2+ and Mg2+) and lymphocyte separation medium were obtained from PAA. Flagellin, R848 and Pam3CSK4 were purchased from Enzo Life Science. CpG was from Hycult Biotechnology. Macrophage-activating lipopeptide (MALP-2) was obtained from Imgenex. The purity of TLR agonists was estimated for CpG 85% (HPLC), flagellin 95% (SDS-PAGE), LPS 80%, MALP-2 95% (HPLC), poly(I:C) 99% (thin layer chromatography) and LTA 97% according to the manufacturer's statement. All agonists, except LPS, were endotoxin free.
Cell Culture
Human monocyte-derived macrophages (hMDMs) were differentiated from peripheral blood mononuclear cells (PBMCs), as described previously [12]. Blood was obtained from the Red Cross, Krakow, Poland. The Red Cross de-identified blood materials as appropriate for human subjects confidentiality assurance. Thus, the current manuscript adheres to appropriate exclusions from human subjects approval. Briefly, PBMCs were isolated from human blood using a lymphocyte separation medium density gradient yielding the fraction highly enriched in monocytes (90% CD14 positive), as described previously [12]. Cells were plated at 3 × 106/well in 24-well plates (Sarstedt) in RPMI-1640 supplemented with 2 mML-glutamine, 50 µg/ml gentamycin and 10% autological human serum. After 24 h, non-adherent PBMCs were removed by washing with complete medium, and adherent cells were cultured in this medium for 7 days with fresh medium changed every 2 days.
Incubation of hMDMs with TLR Agonists
hMDMs were incubated for various lengths of time in 0.5% human serum RPMI with PBS or the following TLR agonists: LPS (100 ng/ml), LTA (10 µg/ml), R848 (10 µM), CpG-DNA (12 µM), Pam3CSK4 (1 µg/ml), MALP-2 (10 ng/ml), poly(I:C) (10 µg/ml) and flagellin (100 ng/ml).
Bacterial, Fungus and Virus Preparation
S. aureus strains ATCC 29213 and Newman (kindly provided by Dr. T. Foster, Trinity College, Dublin, Irland) and E. coli strain (ATCC 25922) were inoculated from stocks into 10 ml tryptic soy broth media and grown to the stationary growth phase at 37°C overnight under constant rotation (180 rpm). C. albicans (ATCC 10231) were cultured in pepton-yeast extract medium with 4% of glucose at 25°C overnight under constant rotation (180 rpm). Prior to each inoculation, the bacterial and fungus cells were collected by centrifugation (5,000 g, 5 min), washed with PBS and resuspended in PBS to the desired OD600 nm. For some experiments, numbers of vital staphylococci in samples were verified by plating dilutions on agar plates and counting colonies to determine colony-forming units (CFU)/ml according to a previously published protocol [13]. Adenovirus type 1 stock (kindly provided by Dr. K. Pyrć, Jagiellonian University, Krakow, Poland) was generated in A549 cells. Cells were lysed at 3 days after infection by three freeze-thaw cycles. A control A549 cell lysate from mock-infected cells was prepared in the same manner as the virus stocks.
Short-Term Infection of Macrophages
Macrophage infection was performed using S. aureus ATCC 29213 (multiplicity of infection, MOI 1:5), E. coli (MOI 1:5), adenovirus 1 (2,000 TCID50) and C. albicans (MOI 1:20). Phagocytosis assays were carried out for 2 h at 37°C at an indicated MOI. After that cells were rinsed 4 times with ice-cold PBS and infected cells were cultured in medium containing gentamycin (50 µg/ml) for an additional 2 h.
Long-Term Infection of Macrophages
Two models of long-term infection of macrophages were performed. In the first model, we used S. aureus ATCC 29213 or E. coli at the low MOI 1:5, and the level of MCPIP1 mRNA was qualitatively determined in infected cells. In the second model, hMDMs were infected with the Newman strain at the high MOI of 1:50, and we analyzed the MCPIP1 expression until S. aureus escaped from macrophages several days after infection. In both models, 2 h after phagocytosis, cells were rinsed 4 times with ice-cold PBS. Any remaining non-phagocytized bacteria were killed by culturing in medium containing gentamycin (50 µg/ml) for 24 h. The medium was then replaced with fresh media without antibiotics, and cultures were maintained for the desired time.
Quantitative Reverse Transcription PCR
Total cellular RNA was extracted from cultured hMDMs using an RNeasy Mini Kit (Qiagen), according to the manufacturer's instructions. RNA samples were DNase treated and cDNA was prepared by reverse transcription (RT) using the RevertAid™ First Strand cDNA Synthesis Kit (Fermentas). Five hundred nanograms of RNA from each sample was used for cDNA synthesis reaction with oligo(dT) primers, according to the manufacturer's instructions. Quantitative PCR (qPCR) reaction was performed with a SYBR Green method in a reaction volume of 20 µl, containing 1 µl of cDNA sample, 0.5 µM of each primer and 1 × SYBR Green JumpStart Taq Ready Mix. qRT-PCR forward and reverse primers for MCPIP1(ZC3H12A) and for housekeeping EF2 gene (used for normalization) were used previously [4]. After 5 min of initial denaturation at 95°C, reactions were carried out for 40 cycles at the given conditions: denaturation, 95°C, 30 s; annealing, 62°C, 30 s; extension, 72°C, 30 s, followed by a final elongation step at 72°C for 10 min. All the reactions were performed in duplicates. Means for threshold cycle (Ct) values were calculated and analyzed using the ‘delta-delta Ct’ quantification method [14]. Routinely, for the evaluation of quality of qRT-PCR reactions, samples were resolved on non-denaturing 1.5% agarose gels and visualized by staining with ethidium bromide.
Cytokine Assay
Five hundred microliters of cell culture supernatants were collected and stored at −80°C until analysis. The level of IL-6 and/or interferon (IFN)-γ was determined by using commercially available ELISA kits, according to the manufacturer's instructions (BD Bioscience).
Protein Isolation and Immunoblotting
Whole cellular extracts from control and stimulated cells were prepared using 100 µl of RIPA-lysis buffer (0.25% Na-deoxycholate, 0.5% Nonidet P-40, 0.05% SDS, protease inhibitor cocktail, 2.5 mM EDTA in PBS) and stored at −20°C. Equal amounts of protein (40 µg/well) were separated using SDS-PAGE 10% gels and electrotransferred onto nitrocellulose membranes (nitrocellulose BioRad) in buffer composed of 25 mM Tris, 0.2 M glycine, 20% methanol (30 V, overnight). Non-specific binding sites were blocked with 3% BSA in TTBS buffer (20 mM Tris, 0.5 M NaCl, pH 7.5 with 0.05% Tween-20) for 1 h, followed by 1– 2 h incubation with the relevant primary antibody: 1,000-fold diluted rabbit anti-MCPIP1 (Genetex) or 10,000-fold diluted mouse anti-β-actin (BD Bioscience). Membranes were washed extensively in TTBS buffer and incubated with secondary horseradish peroxidase-conjugated antibodies, 10,000-fold diluted donkey anti-rabbit IgG or 20,000-fold diluted sheep anti-mouse IgG, for 1 h in TTBS buffer containing 1% BSA. Membranes were washed (4 × 15 min) in TTBS buffer and blots were developed using ECL detection (Western blotting detection reagents; Amersham Biosciences).
Densitometric Analyses
Densitometric analyses of Western blots were performed using Kodak Digital Software. Results are presented as increases over levels in non-stimulated cells.
Statistics
All experiments were performed at least in triplicate, and results are expressed as means ± SD. Results were analyzed for statistical significance using the non-parametric Student t test. Differences were considered significant when p < 0.05.
Results
Effects of TLR Agonists on MCPIP1 Expression in Macrophages
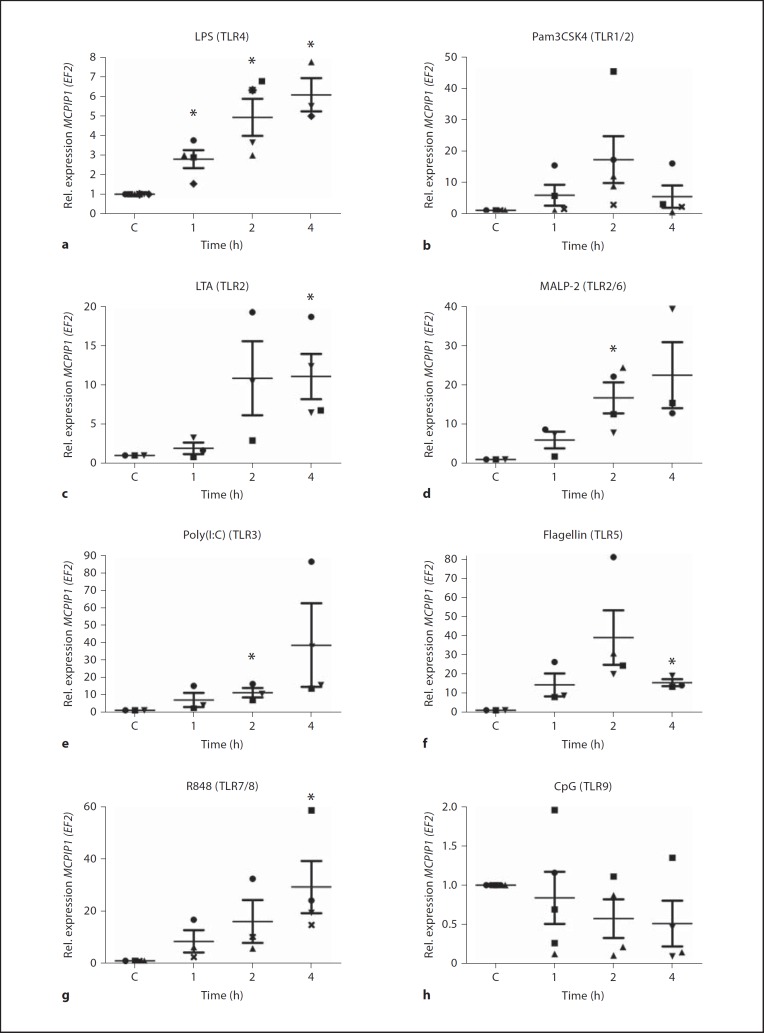
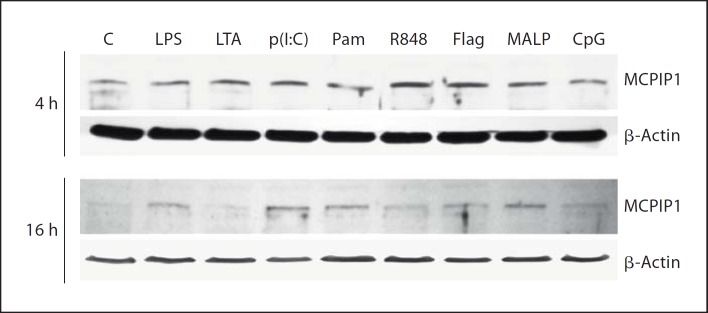
A strong increase in MCPIP1 expression in macrophage cell lines (Raw 264.7 and THP-1) in response to treatment with LPS was recently described [6]. To establish an equivalent and relevant human primary cell model for the evaluation of MCPIP1 expression upon activation of other TLRs, hMDMs were stimulated with 100 ng/ml LPS. qRT-PCR analysis (fig. 1a) revealed increased MCPIP1 expression after 1 h of LPS treatment, which reached its maximum at 4 h (6.09 ± 0.84 fold above the control level). Since each TLR recognizes a distinct pathogen-associated molecular pattern (PAMP), primary macrophages were challenged with: LTA from S. aureus, synthetic lipoprotein (palmitoylated N-acyl-S-diacylglycerylcysteine, Pam3CSK4), MALP-2, dsRNA [poly(I:C)], bacterial flagellin, imidazoquinoline pharmaceutical compound (R848), unmethylated CpG-DNA, which are TLR2, TLR1/2 heterodimer, TLR2/6 heterodimer, TLR3, TLR5, TLR7/8 heterodimer and TLR9-specific stimuli, respectively. The analysis performed by RT-PCR revealed differential upregulation of MCPIP1 at the level of transcription (fig. 1b, c, d, e, f, g, h). Although initial gene induction was observed 1 h after treatment, with the majority of TLR ligands, the timing of maximal MCPIP1 expression level varied from agonist to agonist. A similar pattern was observed for Pam3CSK4, LTA and flagellin (fig. 1b, c, f), where maximal MCPIP1 expression peaked at 17.23 ± 7.4, 10.89 ± 4.7 and 39.10 ± 14.1 (mean value ± SD), respectively, after 2 h. In the case of stimulation with LTA, the maximal expression was sustained up to 4 h. Conversely, the level of MCPIP1 mRNA decreased 4 h after stimulation with Pam3CSK4 and flagellin. The second profile with the highest induction of the MCPIP1 gene after 4 h was observed when cells were treated with MALP-2, poly(I:C) and R848. These TLR2/6-, TLR3- and TLR7/8-specific agonists caused a fold increase in MCPIP1 transcript of 22.57 ± 8.4 (MALP-2), 38.5 ± 23.9 [poly(I:C)] and 31.2 ± 14.2 (R848), respectively, above the basic mRNA level in unstimulated cells (fig. 1d, e, g). Interestingly, MCPIP1 expression was not changed upon CpG stimulation (fig. 1h). Furthermore, the MCPIP1 protein level determined by specific antibody 4 and 16 h after agonist stimulation resembled the mRNA expression profile (fig. 2). The strongest increase was observed 4 h after cells were treated with R848 and flagellin (5.33 ± 3.1 and 5.98 ± 3.2 fold increase above the control level, respectively). However, the profile of the MCPIP1 protein level varied 16 h after cell stimulation with agonists. At that time, the most potent MCPIP1 inducers were LPS, poly(I:C), Pam3CSK4 and MALP-2. Interestingly, CpG increased MCPIP1 slightly (1.69 ± 0.51 fold) but only 4 h after macrophage stimulation. Collectively, these results clearly show that all TLR agonists, except CpG, induce strong MCPIP1 expression in human macrophages.
Fig. 1.
Time-dependent modulation of MCPIP1 expression by TLR agonists. hMDMs were incubated with PBS or TLR ligands: LPS, 100 ng/ml(a);Pam3CSK4, 1 µg/ml (b); LTA, 10 µg/ml (c); MALP-2, 10 ng/ml (d); poly(I:C), 10 µg/ml (e); flagellin, 100 ng/ml (f); R848, 10 µM (g), and CpG-DNA, 12 µM (h). After the indicated time, total RNA was isolated and RT-PCR was performed to estimate the level of the MCPIP1 transcript. Data represent the mean values calculated from the results of 3–5 independent experiments using hMDMs derived from different donors. Rel. = Relative; C = control. Bars represent the mean relative expression ± SD. * p < 0.05.
Fig. 2.
The changes in MCPIP1 protein levels upon hMDMs treatment with TLR agonists. After the stimulation with TLR agonists, cells were cultured for 4 and 16 h, and then protein fractions were prepared as described in Materials and Methods. A representative immunoblot out of 3 separate experiments performed on hMDMs derived from different donors is shown. MCPIP1 was visualized by immunoblot using anti-MCPIP1-specific antibodies. C = Control; p(I:C) = poly(I:C); Pam = Pam3CSK4; Flag = flagellin.
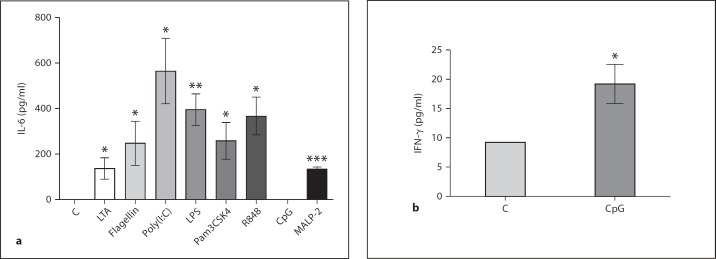
To validate the macrophage activation by the tested TLR agonists, the level of secreted inflammatory mediators was assessed. IL-6 and/or IFN-γ were measured in cell culture medium after stimulation with TLR ligands. Results presented in figure 3 clearly indicate macrophage activation with all ligands tested.
Fig. 3.
Activation of macrophages by TLR agonists determined by the level of proinflammatory cytokine secretion. hMDMs were untreated or exposed to TLR agonists. At 4 (a) or 24 h (b) after ligand distribution, culture supernatants were harvested and assayed for the presence of IL-6 (a) or IFN-γ (b), respectively, by ELISA, as described in Materials and Methods. Bars represent the mean concentration of secreted cytokines ± SD. * p < 0.05; ** p < 0.01; *** p < 0.001.
Induction of MCPIP1 during Short-Term Macrophage Infection
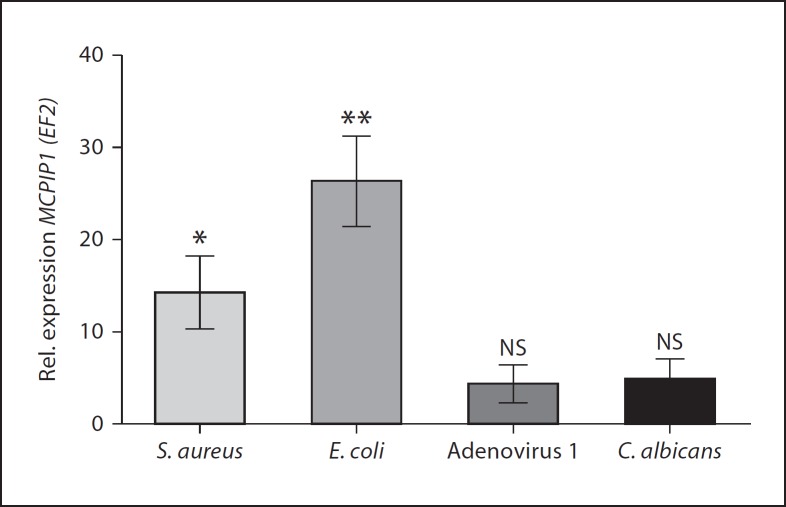
We next investigated whether MCPIP1 induction in hMDMs occurs during infection with live carriers of PAMPs. Therefore, macrophages were infected with Gram-positive (S. aureus) and Gram-negative (E. coli) bacteria, DNA virus (adenovirus type 1) and fungus (C. albicans). The results shown in figure 4 confirmed the observation with pure PAMPs, since in comparison to non-infected cells, each pathogen potently induced MCPIP1 expression within 4 h after infection. The strongest effect was observed during S. aureus and E. coli infection (14.28 ± 3.9 and 26.4 ± 4.8 fold stimulation, respectively). The bacterial intracellular load in infected hMDMs was estimated for S. aureus and E. coli at 192,000 ± 3,421 and 451 ± 89 CFU per 300,000 macrophages, respectively. C. albicans and adenovirus type 1 induced MCPIP1 at a significantly lower level (4.8 ± 2.29 and 4.36 ± 2.08 fold, respectively).
Fig. 4.
The expression of MCPIP1 in hMDMs infected with different pathogens. MCPIP1 expression in macrophages non-infected and infected with S. aureus (ATCC 29213), E. coli, C. albicans or adenovirus type 1 for 4 h was monitored by qRT-PCR. Data represent the mean values calculated from the results of 3 independent experiments using hMDMs derived from different donors. Rel. = Relative; NS = not significant. Bars represent the mean relative expression ± SD. * p < 0.05; ** p < 0.01.
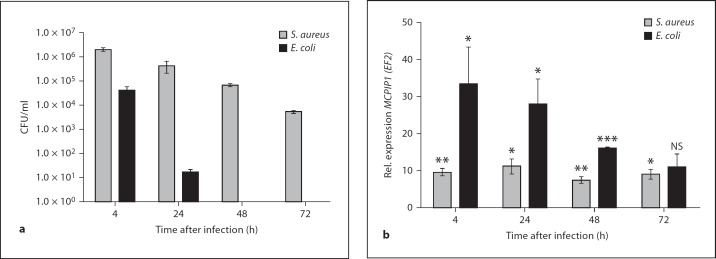
MCPIP1 Regulation during Long-Term Bacterial Infection
A lot of attention has been focused on the role of MCPIP1 in the immediate cellular response to proinflammatory stimulation, but little is known about its functioning in long-term responses induced by pathogens. We previously described that phagocytosis of S. aureus by hMDMs led to prolonged, silent intracellular survival (up to 7 days) of this pathogen in phagocytes without affecting the viability of host cells [13]. In addition, global analysis of the changes in gene expression in S. aureus-infected hMDMs revealed a significant increase in the expression of the MCPIP1 gene in comparison to 18S rRNA after 8 h (3.0 fold), 24 h (3.4 fold) and 48 h after phagocytosis (2.4 fold) [12]. Therefore, the hMDMs infection model was applied to explore MCPIP1 expression in response to intracellular persistence of the pathogen established by infection of hMDMs with a low dose of S. aureus ATCC 29213 (MOI 1:5). For comparison, E. coli (ATCC 25922)-infected hMDMs (MOI 1:5) were used, since this strain is efficiently killed inside phagolysosomes shortly after phagocytosis (fig. 5a). MCPIP1 expression was determined by quantitative real-time PCR at different time points after infection (fig. 5b). After rapid and strong upregulation of MCPIP1 upon infection with both pathogens, a significant decrease in expression was observed only in E. coli infected macrophages (fig. 5b). In contrast, the elevated expression of MCPIP1 was sustained or only slightly decreased during the course of infection, yet at 72 h, it was still 10-fold higher than in mock-infected control cells (fig. 5b). Since we have shown that intracellular survival of S. aureus stimulated a sustained high level of the MCPIP1 mRNA in the infected cells, it was tempting to determine whether S. aureus escaping from macrophages may also modulate MCPIP1 expression. Thus, the second model of long-term staphylococci infection was used. This model allows the observation of pathogen escape from the host cell within several days after infection. Macrophages were infected with a high dose (MOI 1:50) of S. aureus Newman strain. At consecutive days after infection, extracellular bacteria (CFU) were enumerated by plating the medium and counting the CFU, and the MCPIP1 transcript level was determined. Interestingly, a dramatic spike in the level of the MCPIP1 transcript was observed coincident with S. aureus escape from macrophages and massive amplification of bacteria in the growth media (table 1). Taken together, these results clearly show that high expression of MCPIP1 is sustained during long-term bacterial infection and implies that MCPIP1 can serve as a highly sensitive marker of intracellular bacterial infection by S. aureus.
Fig. 5.
Comparison of the effect of long-term infection of macrophages with S. aureus and E. coli on MCPIP1 expression. a At consecutive days after infection, cell lysates were collected and plated onto tryptic soy agar for enumeration of intracellular bacteria (CFU). b MCPIP1 expression in infected hMDMs was monitored by qRT-PCR. Rel. = Relative; NS = not significant. Data represent mean values ± SD calculated from the results of a representative experiment. * p < 0.05; ** p < 0.01; *** p < 0.001.
Table 1.
Correlation between MCPIP1 expression and S. aureus escape from macrophages
| Time after infection h |
Donor 1 |
Donor 2 |
Donor 3 |
|||
|---|---|---|---|---|---|---|
| extracellular bacteria, CFU/ml | MCPIP1/EF2 relative expression | extracellular bacteria, CFU/ml | MCPIP1/EF2 relative expression | extracellular bacteria, CFU/ml | MCPIP1/EF2 relative expression | |
| 24 | 0 | 7.71 | 0 | 5.41 | 0 | 4.45 |
| 48 | 0 | 2.59 | 7.4 × 104 | 22.94 | 0 | 4.71 |
| 72 | 0.64 × 106 | 32.64 | ND | ND | 0 | 0.64 |
| 96 | ND | ND | ND | ND | 0 | 0.97 |
| 120 | ND | ND | ND | ND | 9.9 × 105 | 344.4 |
Macrophages were allowed to engulf S. aureus (Newman strain) at an MOI of 50 for 2 h, were washed, and then extracellular bacteria were killed by gentamycin. Macrophages were cultured in media without antibiotics. At consecutive days after infection, media were aspirated and plated onto TSA for enumeration of extracellular bacteria (CFU). Simultaneously, total RNA was isolated and qRT-PCR reaction was performed to estimate the level of the MCPIP1 transcript. The data illustrate results obtained in 3 representative experiments in which intracellular S. aureus escaped from macrophages at different time points after infection. ND = Not defined.
Discussion
MCPIP1 is regulated by a broad range of proinflammatory cytokines (TNF-α, IL-1β, MCP-1) and also by bacterial antigen LPS [2, 4, 7]. Despite the fact that MCPIP1 is described as a TLR-inducible gene [3], the complex modulation of its expression on the mRNA and protein levels upon stimulation with known TLR agonists has never been documented. Here, we described for the first time the broad analysis of MCPIP1 regulation by different TLR ligands. Our results clearly show that with the exception of CpG, all TLR agonists can strongly potentiate MCPIP1 expression at the level of mRNA and produce protein in human macrophages. To minimize the autocrine and paracrine stimulations by secreted cytokines that may contribute to MCPIP1 expression, the duration of the experiments was limited to 4 h. Within 2 h after stimulation, the level of secreted cytokines was negligible; therefore, we suggest that the observed MCPIP1 induction is the direct consequence of TLR activation. The initial induction occurred within an hour of treatment with agonists, but further time-dependent response varied for different pattern recognition receptor agonists. For Pam3CSK4 or flagellin after rapid MCPIP1 induction, the mRNA levels declined in a time-dependent manner, whereas for the rest of the tested ligands, including LPS, the level of the MCPIP1 transcript peaked at 4 h after stimulation. A similar effect was noticed previously in human and mouse macrophages stimulated with LPS (100 ng/ml) [6]. The effect of the sustained high-level MCPIP1 transcript may be a consequence of mRNA stabilization, which has already been documented for some other cytokine gene transcripts after LPS stimulation. Furthermore, the high level of the MCPIP1 protein detected 16 h after stimulation with several TLR agonists may not only confirm the above notion but it also strongly suggests protection of the MCPIP1 protein from intracellular degradation. Taking into account the response to other TLR ligands, it must be underlined that the maximal MCPIP1 expression induced by endotoxin during the time course of experiments was significantly lower (6.09 ± 0.85) compared to the effect induced by other TLR agonists (26.77 ± 5.6). Interestingly, these findings do not correlate with much stronger induction of MCPIP1 by E. coli infections comparing to other pathogens (S. aureus,C. albicans and adenovirus 1). This discrepancy is probably due to the fact that many signaling pathways in addition to TLRs are involved in MCPIP1 induction during infection with live bacteria. Secondly, the difference in induction of MCPIP1 by LPS and other agonists may result from a relatively low dose of endotoxin used in our study. In the case of CpG, which among all tested agonists exerts only slight induction of MCPIP1 at the protein level, such observation can be explained by insufficient expression and, in consequence, by activation of the TLR9 receptor on hMDMs.
The observed difference in MCPIP1 mRNA induction by TLR agonists does not correlate with cellular localization of their receptors [15]. TLRs can be classified into two groups based upon cellular location. TLRs 1, 2, 4, 5 and 6 are found on the cell membrane and can be activated by extracellular PAMPs. By contrast, the presence of TLRs 3, 7, 8 and 9 in membranes of intracellular compartments, such as endosomes and lysosomes, allows detection of nucleic acids (i.e. DNA or RNA) released from viruses or bacteria that are degraded within endosomes and lysosomes inside the cell [15]. MCPIP1 possesses RNase activity that regulates its own transcript [3, 4]. Thus, one may suspect that the decline in MCPIP1 mRNA 4 h after Pam3CSK4 or flagellin stimulation is a consequence of enhanced self-degradation. However, such contention is contradicted by sustained increase in MCPIP1 transcript in the case of hMDM stimulation with R848 and LTA. Therefore, the mechanism of differential activation of MCPIP1 expression needs further examination.
The tissue distribution of MCPIP1 transcript revealed the highest expression of the MCPIP1 gene in leukocytes, suggesting an important role for MCPIP1 in the functioning of the immunological system [4]. In this study, we show that hMDMs present the perfect model to study MCPIP1 regulation upon stimulation of different TLRs, as well as during macrophage interactions with different pathogens. In keeping with the latter suggestion, we showed that MCPIP1 is induced in hMDMs upon the interaction with different live pathogens and that this most likely mimics the situation during the infection process in vivo. Much attention is paid to rapid, short-term upregulation of MCPIP1 [3], whereas little is known about long-term effects. Macrophages play a vital role in host defense against invading pathogens; however, they can provide a safe niche for silent intracellular survival of bacteria as illustrated by S. aureus [13]. Using hMDMs from selected donors, we documented sustained MCPIP1 expression during prolonged S. aureus persistence in macrophages over the course of several days. Since MCPIP1 negatively controls oxidative killing mechanisms and directs macrophages towards the anti-inflammatory phenotype, one may suspect that sustained MCPIP1 expression during infection would support prolonged intracellular survival of S. aureus in cells with compromised antibacterial activity. In contrast to this Gram-positive pathogen, the infection with E. coli leads to stronger upregulation of MCPIP1 shortly after pathogen engulfment, but the level of MCPIP1 decreases parallel to pathogen eradication. The initial strong induction of MCPIP1 upon infection with E. coli may represent a defense strategy against invading pathogens, since MCPIP1 stimulates expression of a variety of proapoptotic genes. It should be underlined that at equivalent MOI, E. coli is more potent in inducing cell death than S. aureus (online suppl. fig. 1, see www.karger.com?doi=10.1159/000339826). The decline in MCPIP1 mRNA level in E. coli infected macrophages within days after infection may suggest the silencing of inflammatory reaction in response to bacterial eradication. Therefore, we propose that depending on the pathogen, the precise regulation and the level of MCPIP1 during infection can be beneficial for either the host or an invader.
The regulation of macrophage activation in response to LPS is suggested as the main MCPIP1 function. However, new data suggest that this protein can play a role in protection against overstimulation of the innate immune response by other TLR agonists. Matsushita et al. [3] demonstrated, in macrophages from Zc3h12a(–/–) [MCPIP (–/–)] mice, a highly increased production of IL-6 and IL-12p40 in response to poly(I:C), LPS, R848 and CpG. However, this study did not distinguish whether the anti-inflammatory effect of MCPIP1 was the consequence of a constitutively high endogenous level of MCPIP1 in leukocytes [4], or whether the observed effect was the result of the MCPIP1 expression induced by TLR signaling. In this context, our data complement this significant observation by showing that instant, strong upregulation of MCPIP1 mRNA expression via signaling through TLRs is essential for the control of macrophage activation.
The majority of the data showing MCPIP1 regulation and function have been obtained using cell lines or murine models. By contrast, all experiments presented here were carried out on human primary macrophages. The study revealed that the level of induction of MCPIP1 by TLR agonists appeared to be donor dependent. Nevertheless, despite the variable degrees of MCPIP1 expression in hMDMs from different donors, the general time-dependent profile of MCPIP1 induction by each agonist was preserved. Significantly, the overall activation of macrophages derived from different donors, monitored by the general profile of cytokine secretion, was at a comparable level. This indicates that the level of MCPIP1 expression in different individuals can vary in a broad range. One might propose that MCPIP1 is a highly sensitive sensor of TLR-signaling agonists, which may differently curb the immune response in different individuals. Therefore, future studies exploring this difference in the context of individual susceptibility to pathogen infection are of great interest.
In summary, our results clearly indicate a role for MCPIP1 as a potent sensor for TLR-dependent activation of the immune system. Future studies will help to elucidate the importance and mechanisms of MCPIP1 involvement in the pathophysiology of human infectious diseases, especially those caused by chronic, persistent infections.
Supplementary Material
Supplementary data
Acknowledgments
This work was supported by grants from the Polish Ministry of Science and Higher Education N N301 050439 (to J.K.) and 776/N-COST/2010/0 (to J.J.). The Faculty of Biochemistry, Biophysics and Biotechnology of the Jagiellonian University is a beneficiary of structural funds from the European Union (POIG.02.01.00-12-064/08).
References
- 1.Medzhitov R, Janeway C., Jr Innate immunity. N Engl J Med. 2000;343:338–344. doi: 10.1056/NEJM200008033430506. [DOI] [PubMed] [Google Scholar]
- 2.Zhou L, Azfer A, Niu J, Graham S, Choudhury M, Adamski FM, Younce C, Binkley PF, Kolattukudy PE. Monocyte chemoattractant protein-1 induces a novel transcription factor that causes cardiac myocyte apoptosis and ventricular dysfunction. Circ Res. 2006;98:1177–1185. doi: 10.1161/01.RES.0000220106.64661.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matsushita K, Takeuchi O, Standley DM, Kumagai Y, Kawagoe T, Miyake T, Satoh T, Kato H, Tsujimura T, Nakamura H, Akira S. Zc3h12a is an RNase essential for controlling immune responses by regulating mRNA decay. Nature. 2009;458:1185–1190. doi: 10.1038/nature07924. [DOI] [PubMed] [Google Scholar]
- 4.Mizgalska D, Wegrzyn P, Murzyn K, Kasza A, Koj A, Jura J, Jarzab B, Jura J. Interleukin-1-inducible MCPIP protein has structural and functional properties of RNase and participates in degradation of IL-1beta mRNA. FEBS J. 2009;276:7386–7399. doi: 10.1111/j.1742-4658.2009.07452.x. [DOI] [PubMed] [Google Scholar]
- 5.Skalniak L, Mizgalska D, Zarebski A, Wyrzykowska P, Koj A, Jura J. Regulatory feedback loop between NF-kappaB and MCP-1-induced protein 1 RNase. FEBS J. 2009;276:5892–5905. doi: 10.1111/j.1742-4658.2009.07273.x. [DOI] [PubMed] [Google Scholar]
- 6.Liang J, Wang J, Azfer A, Song W, Tromp G, Kolattukudy PE, Fu M. A novel CCCH-zinc finger protein family regulates proinflammatory activation of macrophages. J Biol Chem. 2008;283:6337–6346. doi: 10.1074/jbc.M707861200. [DOI] [PubMed] [Google Scholar]
- 7.Liang J, Saad Y, Lei T, Wang J, Qi D, Yang Q, Kolattukudy PE, Fu M. MCP-induced protein 1 deubiquitinates TRAF proteins and negatively regulates JNK and NF-kappaB signaling. J Exp Med. 2010;207:2959–2973. doi: 10.1084/jem.20092641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Suzuki HI, Arase M, Matsuyama H, Choi YL, Ueno T, Mano H, Sugimoto K, Miyazono K. MCPIP1 ribonuclease antagonizes dicer and terminates microRNA biogenesis through precursor microRNA degradation. Mol Cell. 2011;44:424–436. doi: 10.1016/j.molcel.2011.09.012. [DOI] [PubMed] [Google Scholar]
- 9.Jura J, Wegrzyn P, Korostyński M, Guzik K, Oczko-Wojciechowska M, Jarzab M, Kowalska M, Piechota M, Przewłocki R, Koj A. Identification of interleukin-1 and interleukin-6-responsive genes in human monocyte-derived macrophages using microarrays. Biochim Biophys Acta. 2008;1779:383–389. doi: 10.1016/j.bbagrm.2008.04.006. [DOI] [PubMed] [Google Scholar]
- 10.Kasza A, Wyrzykowska P, Horwacik I, Tymoszuk P, Mizgalska D, Palmer K, Rokita H, Sharrocks AD, Jura J. Transcription factors Elk-1 and SRF are engaged in IL1-dependent regulation of ZC3H12A expression. BMC Mol Biol. 2010;11:14. doi: 10.1186/1471-2199-11-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zarember KA, Godowski PJ. Tissue expression of human Toll-like receptors and differential regulation of Toll-like receptor mRNAs in leukocytes in response to microbes, their products, and cytokines. J Immunol. 2002;168:554–561. doi: 10.4049/jimmunol.168.2.554. (erratum published in J Immunol 2002; 169:1136) [DOI] [PubMed] [Google Scholar]
- 12.Koziel J, Maciag-Gudowska A, Mikolajczyk T, Bzowska M, Sturdevant DE, Whitney AR, Shaw LN, DeLeo FR, Potempa J. Phagocytosis of Staphylococcus aureus by macrophages exerts cytoprotective effects manifested by the upregulation of antiapoptotic factors. PLoS One. 2009;4:e5210. doi: 10.1371/journal.pone.0005210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kubica M, Guzik K, Koziel J, Zarebski M, Richter W, Gajkowska B, Golda A, Maciag-Gudowska A, Brix K, Shaw L, Foster T, Potempa J. A potential new pathway for Staphylococcus aureus dissemination: the silent survival of S. aureus phagocytosed by human monocyte-derived macrophages. PLoS One. 2008;3:e1409. doi: 10.1371/journal.pone.0001409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 15.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data