ABSTRACT
Our previous study has demonstrated that knockdown of Grainyhead-like 2(GRHL2) in colorectal cancer (CRC) cells inhibited cell proliferation by targeting ZEB1. This study aimed at researching whether knockdown of GRHL2 promoted CRC progression and metastasis via inducing epithelial-mesenchymal transition (EMT). GRHL2-upregulated SW-620/GRHL2+ and GRHL2-knockdown HCT116/GRHL2-KD, HT29/GRHL2-KD cells and their control cells were generated. The morphological changes after overexpression and knockdown GRHL2 were observed. qRT-PCR, Western blotting, and Immunofluorescence were used to detect EMT markers: E-cadherin, Vimentin, p-catein, ZO-1 and ZEB1 expression. Then, sh-ZEB1 was transfected to GRHL2 knockdown cells to research the relationship between GRHL2 and ZEB1. Transwell and wound healing assays were further performed to detect the impact of GRHL2 on invasion and migration in vitro. CRC cells were injected into mice tail vein to verify the impact of GRHL2 on CRC metastasis. Morphological change of mesenchymal-epithelial transition (MET) could be observed in SW620/GRHL2+ cell. The expression of epithelial markers: E-cadherin, β-catenin, ZO-1 were up-regulated, while mesenchymal markers: Vimentin was decreased. Meanwhile, opposite EMT morphological change could be observed in HCT116/GRHL2-KD cell, accompanied by reverse change of E-cadherin, β-catenin, ZO-1, and Vimentin. The expression level of GRHL2 and ZEB1 was found negative in both SW620/GRHL2+ and HCT116/GRHL2-KD cells. Knockdown of ZEB1 by siRNA in HCT116/GRHL2-KD and HT29/GRHL2-KD could upregulate expression of E-cadherin and GRHL2. GRHL2 knockdown also promoted migration, invasion in vitro and CRC metastasis in mice model. In conclusion, GRHL2/ZEB1 axis inhibits CRC progression and metastasis via oppressing EMT.
KEYWORDS: Grainyhead-like 2, colorectal cancer, epithelial-mesenchymal transition, mesenchymal-epithelial transition, ZEB1
Introduction
Colorectal cancer (CRC) is one of the major leading causes of cancer-related death. The majority of CRC patients are diagnosed at a late stage. Despite the remarkable accomplishments in new therapeutic options, the outcome for CRC patients remains poor, particularly those with metastasis.1
GRHL2 regulates the formation of apical junction complexes by regulating the cis-regulatory elements of the core promoter of CLDN4 and intron 2 of the E-cadherin gene, thus participating in the fractionation of epithelial cells.2 In oral squamous cell carcinoma, GRHL2 plays a transcriptional regulatory role by binding specifically to the promoter region – 49 ~+5 of hTERT; meanwhile, the silence of GRHL2 has no effect on the activity of promoter region with mutation.3 GRHL2 can also regulate the methylation of the promoter region of hTERT by inhibiting the activity of DNA methyltransferase DNMT1. Furthermore, Other members of the grainyhead-like family also play a role in transcriptional regulation. In neuroblastoma, HDAC3 and MYCN colocalized to the GRHL1 promoter and repressed its transcription; meanwhile, GRHL1 regulated 170 genes genome-wide, and most were involved in pathways including nervous system development, proliferation, cell-cell adhesion, cell spreading, and cellular differentiation.4 In the process of epidermal keratinocytes transition, GRHL3 represses the formation of a number of progenitors and non-keratinocyte super-enhancers in differentiating keratinocytes. Hence, chromatin relocates GRHL3 binding and enhancers to regulate both the irreversible commitment of progenitor keratinocytes to differentiation and their reversible transition to migration.5 Our previous study has demonstrated that GRHL2 was over-expressed in CRC tissues and positively correlated with tumor size and TNM stage. Kaplan-Meier analysis showed that GRHL2 was an independent prognostic factor for both overall survival and recurrence-free survival. Ectopic over-expression of GRHL2 in CRC cell line HT29 and SW620 induced an increase of cellular proliferation in vitro and promoting tumor growth in vivo. The acquisition of GRHL2 regulated cell cycle and modulates the expression of proliferation proteins p21, p27, cyclin A and cyclin D1.6 We had also identified downregulation of GRHL2 inhibits the proliferation of colorectal cancer cells by targeting ZEB1.7
Epithelial-mesenchymal transition (EMT) refers to the transformation of epithelial cells into mesenchymal cells under specific physiological and pathological conditions.8 The concept of EMT was first proposed in the field of embryonic development. Since then, more and more studies on EMT have been carried out, involving different life phenomena and pathological processes.9 During EMT, intercellular junctions and cell polarity disappear, epithelial markers are down-regulated, mesenchymal phenotypes and related markers are gradually up-regulated, biological behavior of cells is also changed, which is characterized by enhanced migration and invasion ability.10 EMT is closely related to tumor invasion and metastasis and plays an important role in invasion and distant metastasis of many cancers. At the same time, under certain conditions, tumor cells with mesenchymal phenotype can be transformed into tumor cells with epithelial phenotype, that is mesenchymal epithelial transformation (MET).11
The important sign of EMT is the loss of epithelial marker protein (E-cadherin) and the acquisition of interstitial marker proteins (Vimentin, N-cadherin, etc.). The molecular mechanism of EMT is very complex, and many molecules and signal pathways are involved in it. Several key transcription factors can directly act on the promoter of E-cadherin and inhibit its transcription. These transcription factors include Zinc finger protein family ZEB (ZEB1, ZEB2), Snail family (Snaill, Snai12, Snai13), bHLH factors Twist family (Twisty Twist 2) and others. Among them, ZEB1 can directly inhibit the expression of E-cadherin by binding its zinc finger structure to E-box in the promoter region of the E-cadherin gene, thus initiates the EMT process.12
In this research, we engaged in informing regulatory network of GRHL2/ZEB1/E-cadherin in colorectal cancer.
Results
GRHL2 changed stable cell lines were successfully generated
GRHL2 was significantly upregulated in SW620/GRHL2 compare with SW620/Vector and SW620/Parental. Oppositely, GRHL2 was significantly downregulated in HCT116/GRHL2-KD and HT29/GRHL2-KD (Figure 1).
Figure 1.
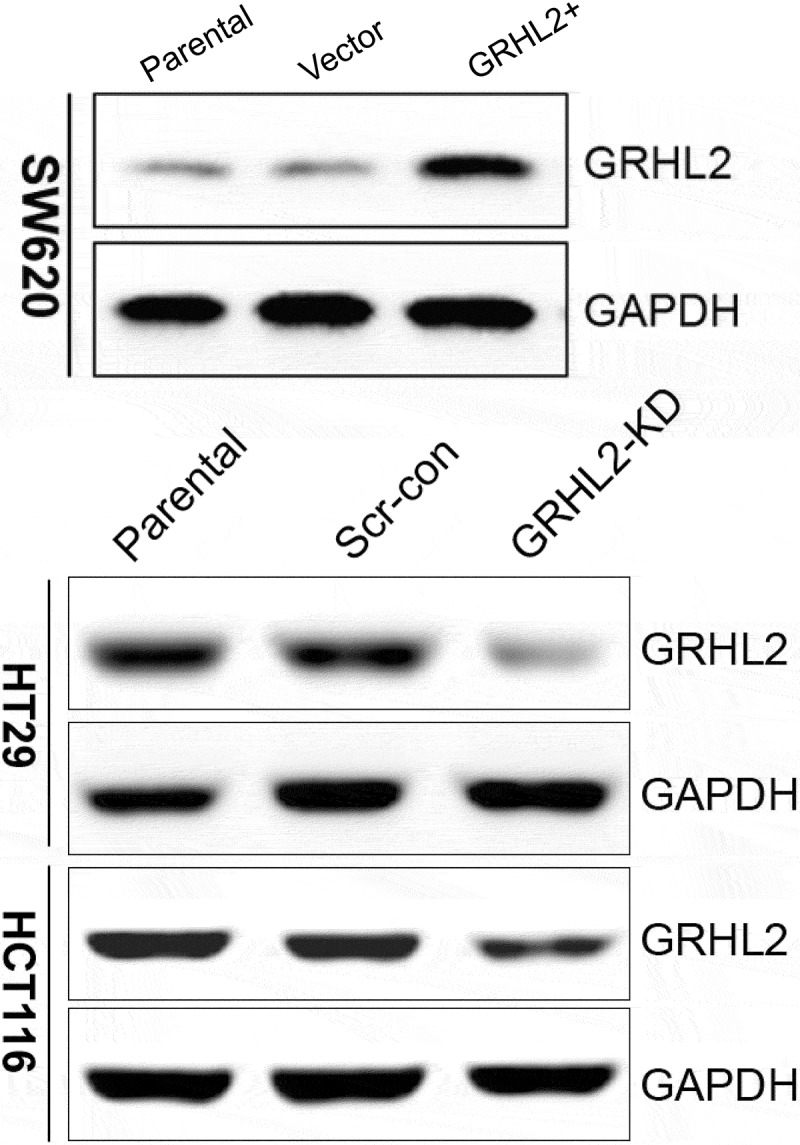
GRHL2 detected in CRC cells by Western blotting.
Overexpression of GRHL2 in SW620/GRHL2+ induced MET
Compared with SW620/Vector cells transfected with blank vector, SW620/GRHL2+ cells were found to have morphological changes (Figure 2A). SW620/GRHL2+ cells restored to an epithelial-like shape, showed an increased tendency to change from a fibroblast-like shape and a scattered growth pattern to growing as even more tightly packed clusters of cells. qRT-PCR (Figure 2B), Western blotting (Figure 2C) and IF (Figure 2D) detect of EMT marker expression revealed a striking down-regulation of the mesenchymal marker (Vimentin) and an up-regulation of epithelial markers: E-cadherin, β-catenin, ZO-1 compared with SW620/Vector and SW620/Parental
Figure 2.
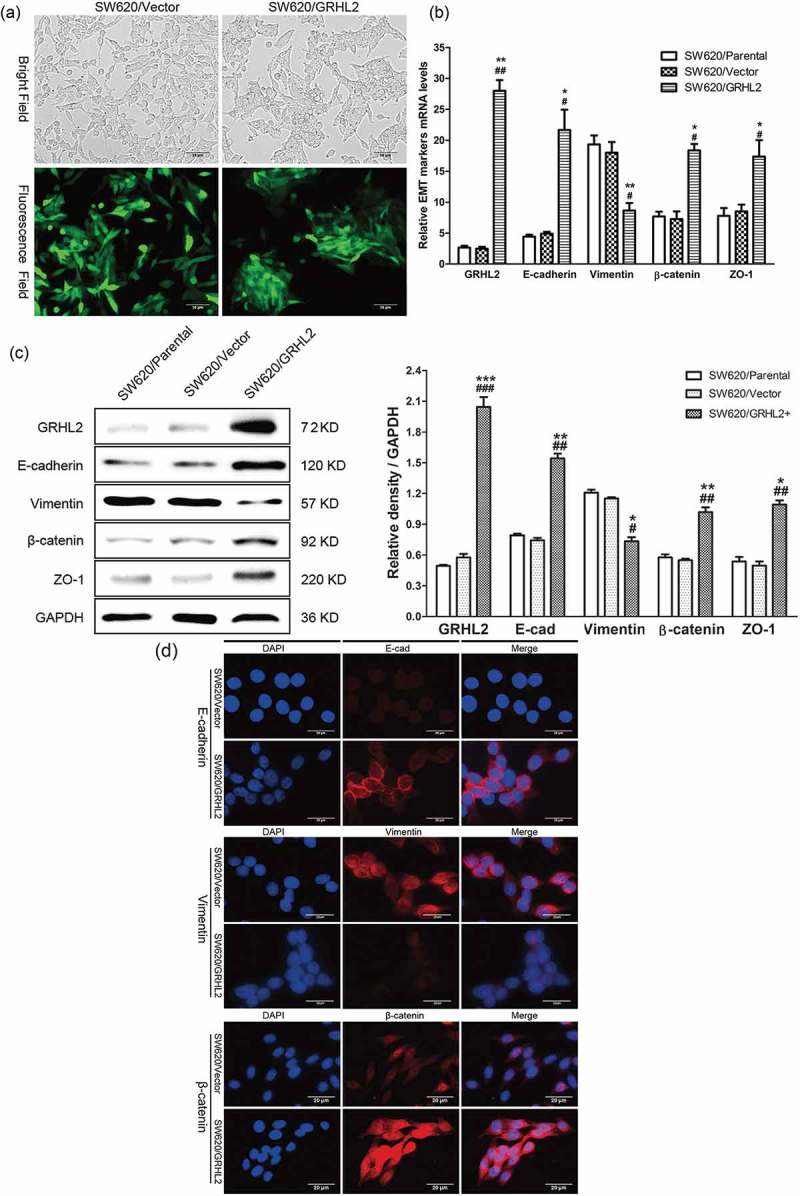
A. Overexpression of GRHL2 induced SW620 cells to undergo a MET-like phenotypical change. B. qRT-PCR analysis of EMT marker gene expression in GRHL2-overexpression cells. C. WB analysis of EMT marker proteins in GRHL2-overexpression cells. D. analysis of EMT marker proteins in GRHL2-overexpression cells. *relative to SW620/Vector control *p < 0.05, **p < 0.01, ***p < 0.001; #relative to SW620/Parental control #p < 0.05,##p < 0.01.
GRHL2 knockdown in HCT116/GRHL-KD induced EMT
HCT116/GRHL2-KD showed an increased tendency to change from tightly packed clusters of cells to a fibroblast-like shape and a scattered growth pattern loss of cell-cell contacts compared with HCT116/Scr-con (Figure 3A). EMT markers expression was detected by qRT-PCR (Figure 3B), Western blotting (Figure 3C) and immunofluorescence (Figure 3D). EMT markers revealed a striking upregulation of selected mesenchymal protein (Vimentin) and a concomitant downregulation of epithelial markers: E-cadherin, β-catenin, ZO-1.
Figure 3.
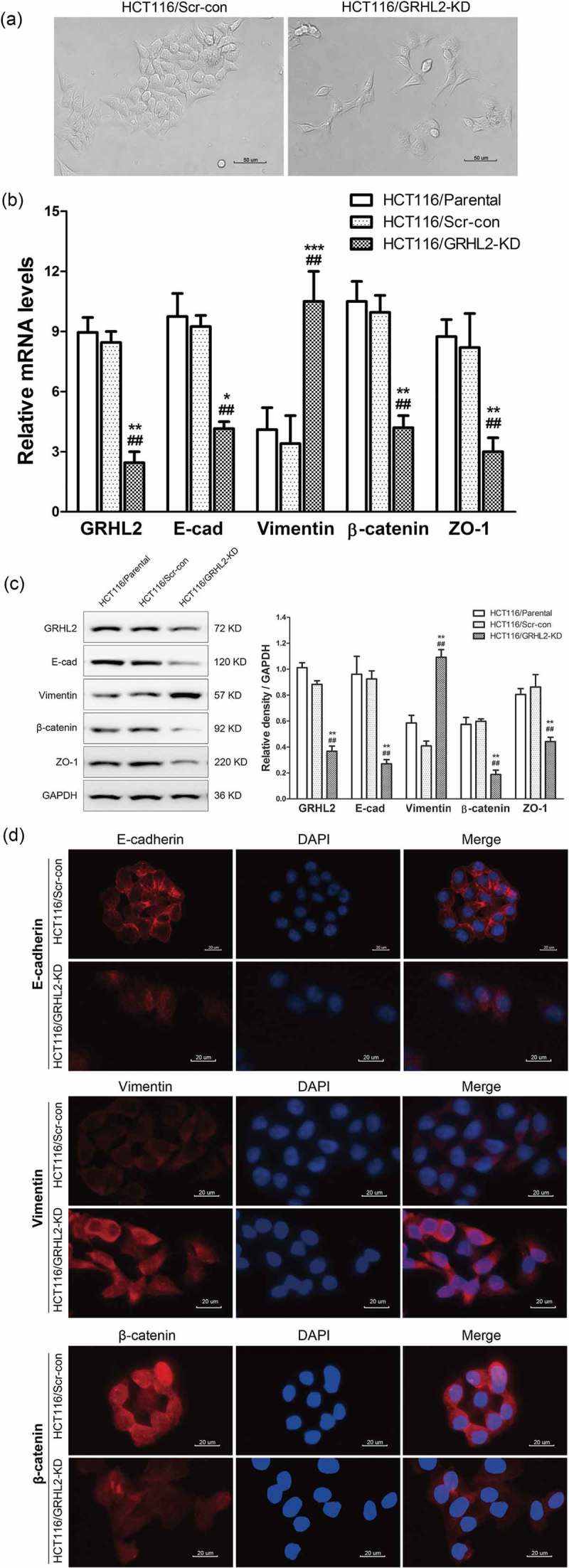
A. GRHL2 knockdown induced HCT116 EMT morphology change. B. qRT-PCR analysis of EMT marker gene expression in GRHL2 knockdown cells. C. WB analysis of EMT marker proteins in GRHL2-knockdown cells. D. IF analysis of EMT marker proteins in GRHL2-knockdown cells. *relative to HCT116/Scr-con control *p < 0.05, **p < 0.01, ***p < 0.001; #relative to HCT116/Parental control #p < 0.05, ##p < 0.01.
Negative expression level of GRHL2 and ZEB1 in CRC cells
ZEB1 protein and transcriptional expression in SW620/GRHL2+, SW620/Vector, HCT116/GRHL2-KD, HCT116/Scr-con was detected by qRT-PCR (Figure 4A), IF (Figure 4B) and Western blotting (Figure 4C). ZEB1was decreased in SW620/GRHL2+ compared with SW620/Vector; oppositely, ZEB1 was increased in HCT116/GRHL2-KD compared with HCT116/Scr-con.
Figure 4.
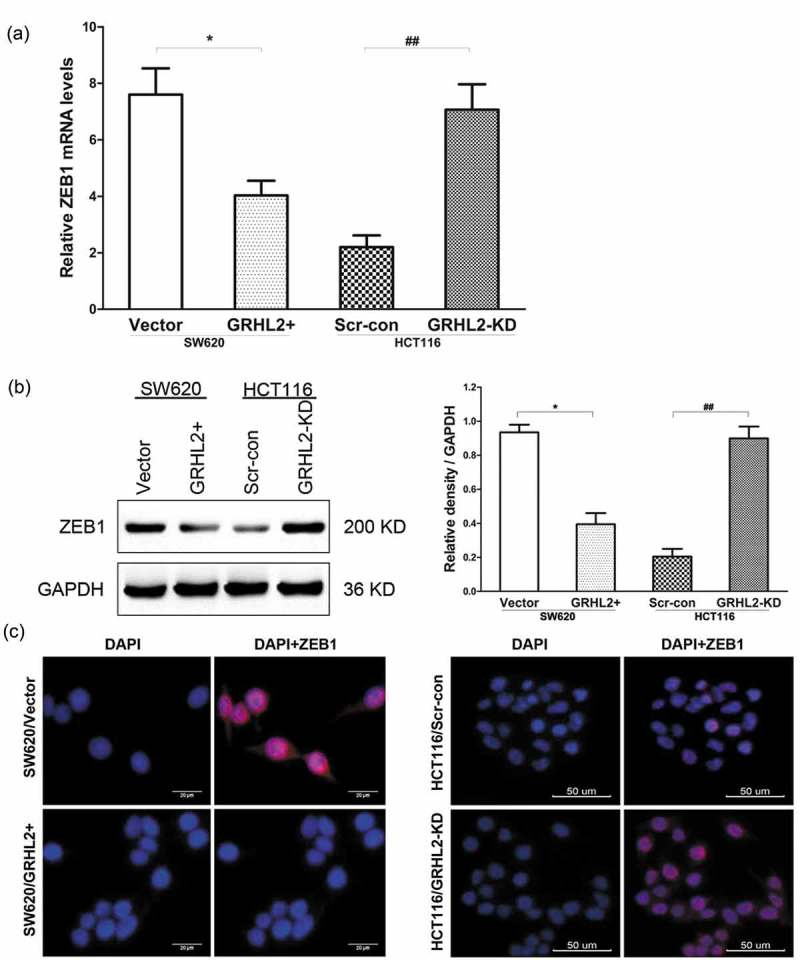
A. ZEB1 expression detected by qRT-PCR. B. ZEB1 expression detected by WB. C. A. ZEB1 expression detected by IF. *relative to SW620/Vector control, *p < 0.05; #relative to HCT 116/Scr-con control, ##p < 0.01.
Knockdown of ZEB1 upregulated expression of E-cadherin and GRHL2
Transient knockdown of ZEB1 using shZEB1 resulted in decreased ZEB1 expression and increased GRHL2, E-cadherin expression in GRHL2-KD HT29 and HCT116 cells at transcriptional (Figure 5A) and protein level (Figure 5B).
Figure 5.
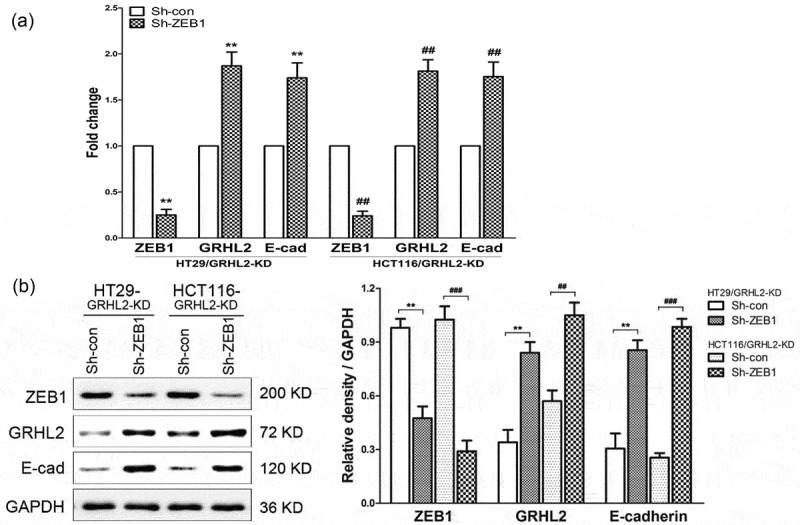
A. qRT-PCR analysis of ZEB1,GRHL2 and E-cadherin at transcriptional expression. B. WB analysis of ZEB1,GRHL2 and E-cadherin at protein expression. *relative to HT29/Sh-con control, *p < 0.05, ** p < 0.01, p < 0.001; #relative to HCT116/Sh-con control, #p < 0.05, ##p < 0.01, ###p < 0.001.
GRHL2 knockdown promoted migration and invasion of CRC cells
To confirm that GRHL2 is an important factor for CRC cells migration and invasion, we examined the migration and invasion ability of the cells by wound healing and transwell assay. HCT116/GRHL2-KD migration and invasion were increased compared with HCT116/Scr-con and HCT116/Parental (Figure 6A). Moreover, HCT116/GRHL2-KD and HT29/GRHL2-KD also showed higher wound healing rate compared with HCT116/Scr-con and HT29/Scr-con (Figure 6B).
Figure 6.
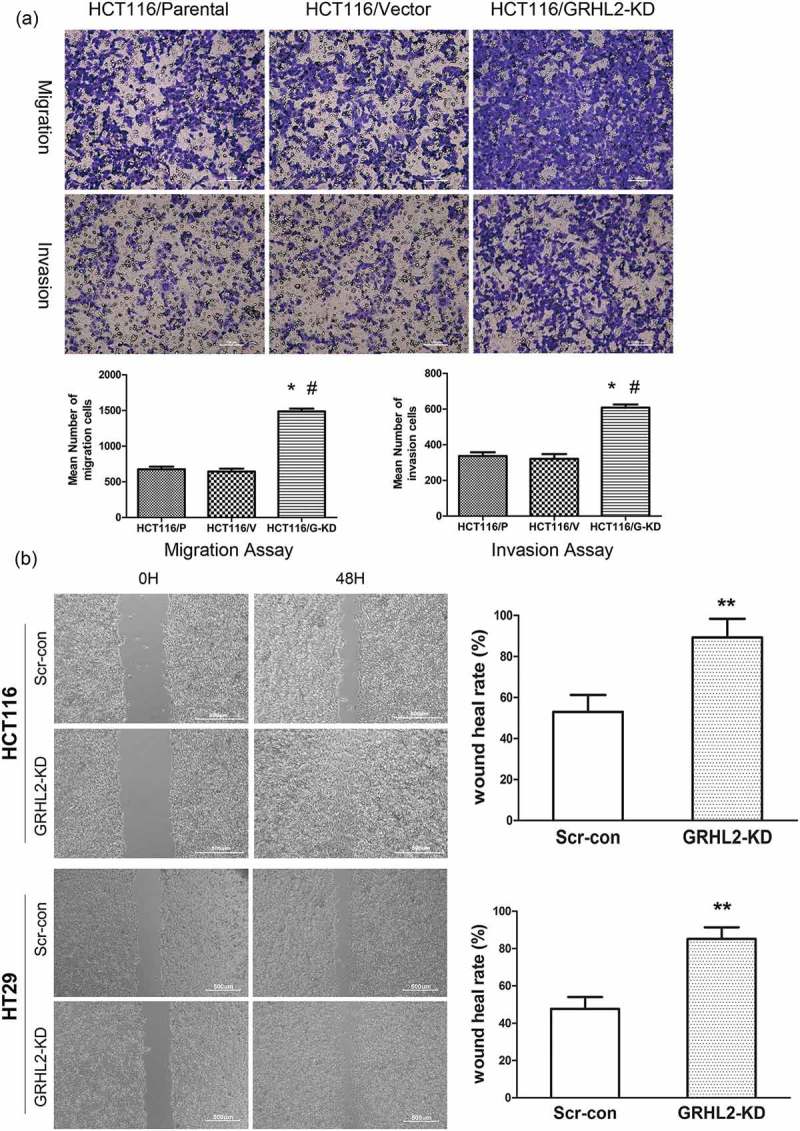
A. GRHL2 knockdown promoted invasion and migration ability of HCT116 cells. B. Wound healing assay demonstrated the promotive effect migration ability of GRHL2-knockdown. *relative to Vector/Scr-con control cells, *p < 0.05, ** p < 0.01, p < 0.001. #relative to Parental control cells, #p < 0.05, ##p < 0.01, ###p < 0.001.
GRHL2 inhibited CRC metastasis in mice model
SW620/GRHL2 +, SW620/Vector, HCT116/GRHL2 – KD and HCT116/Scr – con cells were injected into the tail vein of nude mice to establish the metastasis model. After 5 weeks of injection, most mice began to lose weight and one mice was found multiple subcutaneous metastases in both SW620/Vector and HCT116/GRHL2-KD group (Figure 7A). After 7 weeks, the mice began to show signs of cachexia, bow-back activity, especially in HCT116/GRHL2-KD group, the mice were extremely thin. Two mice of HCT116/GRHL2-KD group died after 7 weeks, and disseminated miliary metastatic nodules were found on their lung surface. After 8 weeks of injection, the remaining mice were sacrificed and dissected routinely. No metastases of liver, kidney, spleen, mesenteric lymph node, and abdominal cavity were found in all groups. Liver slices of each group were stained with HE, and no liver metastases were seen by HE staining (Figure 7B). The lungs of mice in each group showed metastatic foci in varying degrees. Metastatic foci were found in all the lungs of HCT116/GRHL2-KD group, seven in HCT 116/Scr-con group, eight in SW620/Vector and eight in SW620/GRHL2+ groups (Figure 7C). The metastatic foci in the lungs are densely packed with solid masses, nests or cords (Figure 7D). The number of lung metastases in the SW620/GRHL2+ group was significantly lower than that in the SW620/Vector group (3.5 ± 0.5 vs 5.6 ± 0.6, P = 0.016, Figure 7E), whereas the number of lung metastases in the HCT116/GRHL2-KD cell group was significantly higher than that in the HCT116/Scr-con group (9.0 ± 0.8 vs 6.4 ± 0.7, P = 0.043, Figure 7F).
Figure 7.
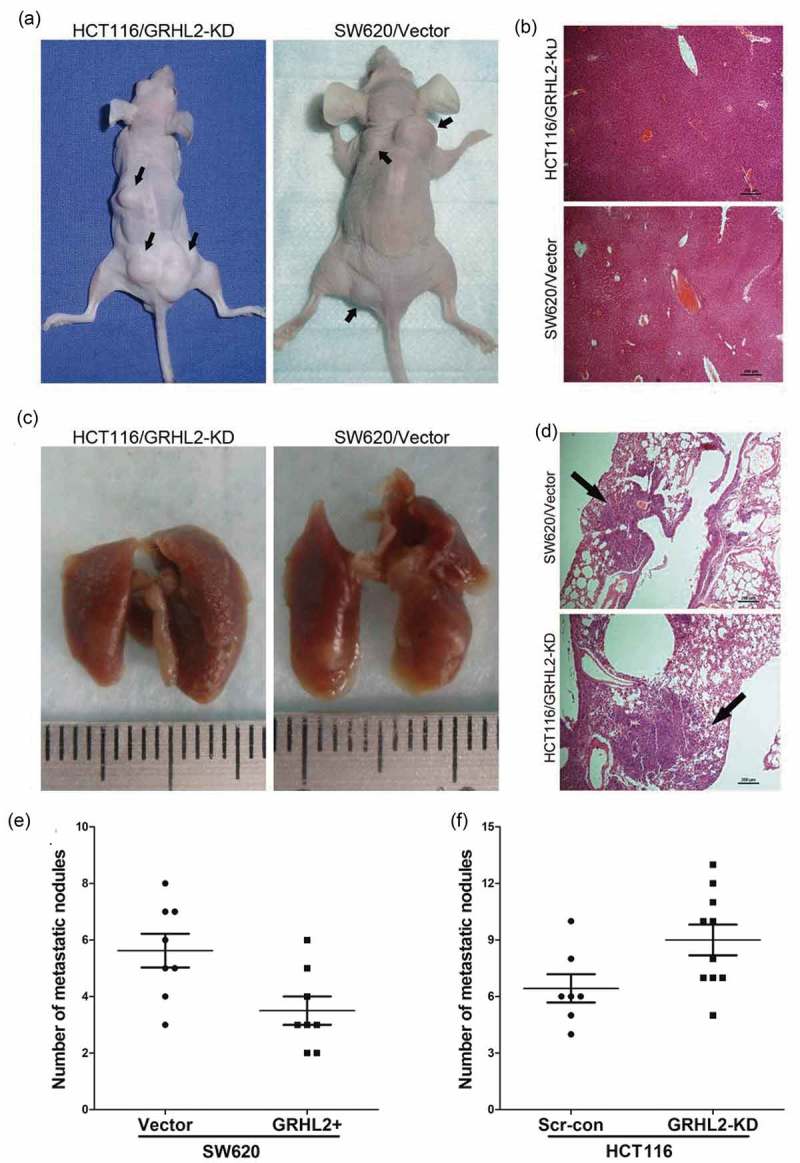
GRHL2 inhibited the metastatic ability of CRC cells in the animal model. A. Two mice suffered from systemic multiple subcutaneous metastases. B. HE staining showed no fiver metastases. C. Multiple metastatic nodules were found on the lung surface of HCT116/GRHL2-KD and SW620/Vector group. D HE staining showed metastatic nodules in the lung of HCT116/GRHL2-KD and SW620/Vector group. The black arrow indicated the pulmonary metastases. E. Compared with the SW620/Vector group the average number of lung metastases in SW620/GRHL2+ group was significantly reduced (3.5 ± 0.5 vs 5.6 ± 0.6, P = 0.016). E. HCT116/GRHL2-KD group had more metastatic nodules than that in HCT116/Scr-con group (9.0 ± 0.8 vs 6.4 ± 0.7, P = 0.043).
GRHL2 had no effect on the EGFR/Ras/Raf/MAPK signaling pathway
EGFR is abnormally activated in many human tumors, promoting the proliferation, invasion, metastasis, and anti-apoptosis of tumor cells. Its abnormal activation mainly activates its downstream Ras/Raf/MAPK pathway. In addition, HCT116 and SW620 are driven by mutant K-RAS signaling; HT29 is driven by a mutant B-RAF. To research GRHL2 effect on EGFR/Ras/Raf/MAPK pathway, Western Blotting was performed. All associated protein expression of EGFR/Ras/Raf/MAPK showed no change in GRHL2 upregulated/knockdown cells compared with control cells. We further detected the Akt pathway, there was also no obvious activity (Figure 8).
Figure 8.

WB analysis of EGFR/Ras/Raf/MAPK and AKT pathways in SW620 and HT29 cells.
Discussion
Isolated single cells or fewer than five undifferentiated cancer cells are often observed at the invasive front of CRC. These cells, called tumor budding, are morphological manifestations of EMT in colorectal cancer and can be observed in 20–40% of the colorectal cancer.13 More and more evidences show that EMT plays an important role in the invasion and metastasis of colorectal cancer. Tumor cells detached from the primary lesion through EMT, then destroyed the basement membrane, invaded blood vessels, survived in the circulatory system, escaped immune surveillance, metastasized to distant organs, and finally formed epithelial metastases through the MET process.
In our study, we overexpressed and knockdown GRHL2 to research GRHL2-regulated EMT in CRC. Overexpression of GRHL2 in SW620 resulted in morphological changes of MET, elevated expression of epithelial markers and decreased expression of interstitial markers. After GRHL2 knockdown of HCT116 cells, EMT morphological changes were observed, accompanied by down-regulation of epithelial markers and up-regulation of interstitial markers. The occurrence of EMT is often accompanied by increased invasiveness and metastasis. These results are in good agreement with our in vitro experiments and tumor metastasis model, suggesting that the role of GRHL2 in the invasion and metastasis of CRC may be partly due to the regulation of EMT.
The important target genes of GRHL2 include ZEB1, which directly inhibits E-cadherin transcription in a variety of tumors.14 GRHL2 has been proved to inhibit ZEB1 expression by directly binding to ZEB1 promoter.15 ZEB1 is considered necessary for tumor cells to maintain interstitial properties and can significantly enhance tumor invasiveness and metastasis rate by inducing the EMT. Several studies have shown that ZEB1 is highly expressed in poorly differentiated malignancies, especially in the invasive frontier, but rarely in normal epithelial tissue or highly differentiated malignancies. ZEB1 can also inhibit the expression of GRHL2 by binding two Z-box elements (located at −129 and −106) and one E-box element (located at −176) to the promoter of GRHL2.15,16 In our study, we informed the negative correlation between GRHL2 and ZEB1 in CRC cell lines. Overexpression of GRHL2 could down-regulate the expression of ZEB1, while knockdown of GRHL2 can up-regulate the expression of ZEB1, which indicates that ZEB1 is a target gene located downstream of GRHL2 in CRC cells. At the same time, we silenced ZEB1 in GRHL2 knockdown cell lines, and we observed an upregulation of GRHL2 and E-cadherin, indicated that GRHL2 is also a target gene for ZEB1. We concluded that there is also negative regulation between ZEB1 and GRHL2 in the EMT process of CRC.
Compared with our previous studies that GRHL2 promotes the proliferation of CRC, GRHL2 plays an inhibitory role in invasion and metastasis. This contradictory result suggests that GRHL2 has diverse biological functions in CRC, similar to other research. GRHL2 also plays a dual regulatory role in the occurrence and development of breast cancer: GRHL2 in the whole breast cancer specimens showed high expression, but in invasion frontier, there is a low expression or expression loss. GRHL2 promotes the proliferation of breast cancer cells by regulating the expression of Erbb3, a member of the epithelial growth factor receptor family; However, GRHL2 also works as an inhibitor of EMT and is involved in the inhibition of invasion and metastasis of breast cancer by regulating ZEB1.15,17 Some growth factors such as TGF-β, FGF, EGF, and IGF activate different intracellular signaling pathways, participate in the process of EMT. Among them, TGF-β is the most thoroughly studied and commonly used EMT inducible factor.18 TGF-β also plays an important role in the regulation of GRHL2, as well as the process of tumorigenesis and development. Similarly, TGF-β works as a tumor suppressor in the early stage of tumorigenesis. But in the late stage of tumorigenesis, TGF-β promotes tumor development by inhibiting apoptosis and promoting invasion and metastasis.19
GRHL2 has been studied in other tumors such as breast cancer in the past, the innovation of our research is slightly inadequate. But we knocked down or overexpressed GRHL2 in three CRC cell lines, sufficiently demonstrated the effect of GRHL2 on EMT in colorectal cancer, which had a certain value.
In summary, our study suggests that GRHL2 plays an important role in EMT of colorectal cancer, and this role depends on the presence of a complex regulatory network between GRHL2/ZEB1/E-cadherin.
Materials and methods
Cell culture
Human colorectal cancer cell lines HCT116, HT29, and SW620 cells were obtained from the American Type Culture Collection (ATCC). HT29, HCT116 cells were grown in McCoy‘s 5A (Corning Cellgro®, Manassas, VA, USA) and SW620 cells were grown in Leibovitz‘s L-15 medium (Corning Cellgro®) medium, supplemented with 10% (v/v) fetal bovine serum (FBS, Corning Cellgro®), penicillin and streptomycin (GIBCO BRL, Gaithersburg, MD, USA). All the cells were cultured at 37℃ in a humidified atmosphere of 5% CO2.
Generation of stable cell line by lentiviral transduction
GRHL2 overexpressed SW620/GRHL2+ and control SW620/Vector, GRHL2 knockdown HCT116/GRHL2-KD, HT29/GRHL2-KD and control HCT116/Scr-con, HT29/Scr-con cell lines were generated in our previous study.6,7 Western blot analysis was used to verify GRHL2 expression.
Western blotting
The total protein was extracted using RIPA lysis buffer (Solarbio, Beijing, China) with 1% phenylmethanesulfonyl fluoride (PMSF). Then, equal amounts (20 μg) of protein determined by BCA protein assay kit (Thermo Fisher Scientific, USA) and separated with 10%SDS-PAGE. The proteins were then transferred to PVDF membranes (0.45 mm, Solarbio, China) and blocked with 5% nonfat milk for 1 h at room temperature. The following rabbit polyclonal antibodies were incubated at 4°C for 12 h: GRHL2, E-cadherin, β-cadherin, Vimentin, ZO-1, ZEB-1, EGFR, p-EGFR, Akt, p-Akt, Erk1/2, p-Erk1/2, K-RAS, B-RAF (1:500, Cell Signaling Technology, USA). GAPDH rabbit polyclonal antibodies (1:4000, Cell Signaling Technology, USA) were used as loading controls and normalization. The secondary antibodies were anti-rabbit antibodies and were conjugated to horseradish peroxidase (HRP) (1:4000, Proteintech, USA). The antibodies were used at a 1:4000 dilution and were incubated for approximately 1 h at room temperature. After rinsing 3 times with PBS for 5 min, the bands were visualized with ECL reagents (Thermo Fisher Scientific, USA) and developed by Omega Lum G (Aplegen, USA).
Immunofluorescence (IF)
Coverslips were laid flat on the bottom of a six-well plate after cleaning, disinfection, and 24 h ultraviolet irradiation. Then, the cells were seeded on the coverslips at a density of 1 × 106 in each well, cultured in an incubator. The coverslips were rinsed with PBS for 5 min 3 times and fixed with 4% paraformaldehyde for 15 min, followed by permeabilization of the cells in 0.2% TritonX-100 for another 20 min. Next, the coverslips were rinsed with PBS again for 5 min 3 times and blocked by incubating cells in 5% BSA for 60 min. Then, the cells were stained with antibodies described in Western blotting method at 1:100 dilution, followed by a 12-h incubation period at 4ºC. After washing the uncombined antibody with PBS for 5 min 3 times, the cells were incubated with Alexa 488-coupled Goat Anti-Rabbit IgG secondary antibody (1:1000, Cell Signaling Technology, USA) for 1 h at 4°C in the darkness. Finally, DAPI was used as a counterstain to label the nuclei. The stained cells were then acquired and photographed with a fluorescent microscope.
RNA extraction and realtime-quantitative PCR
Total RNA was extracted from cells using Trizol (Invitrogen) according to the manufacturer’s protocol. Total RNA (0.5 μg) from each sample was used for first-strand cDNA synthesis using a reverse transcriptional kit (Promega). For RT-PCR, samples were analyzed on a 1.5% agarose gel. The qRT-PCR was performed using cDNA as a template and Universal PCR Master Mix (Applied Biosystems) on an Applied Biosystems 7900HT sequence detection system (Applied Biosystems). Primers used for qRT-PCR analysis were as follows: GRHL2, GGAAATCTAG CCCTGGGTTT G (forward) and TCAGGGAGGA ACGCACTGA (reverse); GAPDH, AAGGTGAAGG TCGGAGTCAA C (forward) and GGGGTCATTG ATGGCAACAA TA (reverse); E-cadherin, CCCACCACGTACAAGGGTC (forward) and CTGGGGTATTGGGGG CATC (reverse); Vimentin, CCGGGAGAAATTGCAGGAGG(forward) and GGTC AAGACGTGCCAGAGAC (reverse); β-catenin, ATGTCCAGCGTTTGGCTGAA (forward) and TGGTCCTCGTCATTTAGCAGTT (reverse); ZO-1, CAACATACAGTGACGCTTCACA (forward) and CACTATTGACGTTTCCCC ACTC (reverse). Relative amount of mRNA was normalized using GAPDH as an endogenous control.
siRNA transfection
Short hairpin RNA against human ZEB1 (Sh-ZEB1) and negative control shRNA (Sh-con) were purchased from GenePharma. HCT116/GRHL2-KD and HCT29/GRHL2-KD cells were seeded in six-well culture plates one day before transfection, and then were transfected transiently using Lipofectamine 2000 (Invitrogen), according to the manufacturer’s instruction. Forty-eight hours after transfection, cells were verified by western blot and used for analysis.
In vitro cell invasion assay and transwell migration assay
For invasion assays, 2 × 105 cells were plated into the top chamber of a transwell chamber (8 μm pore size; Millipore, Billerica, MA, USA) coated with matrigel (BD Bioscience, San Jose, CA, USA). For transwell migration assays, transwell chambers without matrigel were used. Cells were resuspended in 200 μl non-serum culture medium and medium supplemented with 10% FBS was used as a chemoattractant in the lower chamber. The cells were incubated at 37°C for 48 h in a 5% CO2 humidified incubator. Cells that did not invasion through the pores were then cleared softly with a cotton swab, and cells outside the insert were stained in 1% crystal violet for 30 min. Inserts were washed and photographed and stained cells of five random fields were counted. The relative invasion and migration capacity were interpreted as the average number of cells ± SD per field.
Wound healing assay
5 × 105 cells were seeded into 6-well plates and cultured at 37°C for 24 h. We used a 200 μl sterile micropipette tip to scratch the confluent monolayers in a straight line when cells were 80–90% confluent. Then, we washed floating cells with PBS three times and continued to culture the cells after changing the complete medium to serum-free medium. Images of the same wound position were taken after at 0 h and 48 h under a microscope. The migration results were tested by ImageJ software.
In vivo animal metastasis model
male Balb/cnu/nu mice were purchased from Slaccas Animal Co. (Shanghai, China). Animals were kept in the specific pathogen free (SPF) room of Pudong Hospital, Fudan university. All mice were randomly divided into four groups for injection of different cells (SW620/GRHL2+, SW620/Vector, HT119/GRHL2-KD, HT119/Scr-con), 10 in each group. mice were injected with 1 × 106 cells (100μl). The mice were fed to 8 weeks to sacrifice, and the state was observed every day. The lung, liver, and metastases were removed completely and stained with hematoxylin-eosin (HE). All the organizations were observed and photographed under the microscope.
Statistical analysis
A statistical package of IBM SPSS software (version 19.0, Inc., Chicago, IL) was used for statistical analysis. GraphPad Prism (version 7, GraphPad Software, La Jolla, CA, USA) was used to determine the statistical results. All data are expressed as the mean + standard deviation (mean + sd). The statistical analysis of the data from two groups was performed using a t-test. The comparisons of multiple groups were performed by one-way ANOVA and then an LSD-t test. P < 0.05 was considered to be significant.
Funding Statement
This study was funded by Academic Leaders Training Program Supported by Pudong Health Bureau of Shanghai (Grant No. PWRd2016-05) and Natural Science Foundation of Shanghai (Grant No.16411972800).
Availability of data and material
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
Authors‘ contributions
Zhou Yang and Dejun Wu are equally to the implementation of the experiment; Yushen Cheng contributed to data statistics; Yingjun Quan and Zhijun Min are co-corresponding authors that contributed to research design; All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable
Patient consent for publication
Not applicable
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Pancione M, Remo A, Colantuoni V.. Genetic and epigenetic events generate multiple pathways incolorectal cancer progression. Patholog Res Int. 2012;2012:509348. doi: 10.1155/2012/509348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Werth M, Walentin K, Aue A, Schönheit J, Wuebken A, Pode-Shakked N, Vilianovitch L, Erdmann B, Dekel B, Bader M., et al. The transcription factor grainyhead-like 2 regulates the molecular composition of the epithelial apical junctional complex. Development. 2010;137:3835. doi: 10.1242/dev.055483. [DOI] [PubMed] [Google Scholar]
- 3.Kang X, Chen W, Kim RH, Kang MK, Park NH. Regulation of the hTERT promoter activity by MSH2, the hnRNPs K and D, and GRHL2 in human oral squamous cell carcinoma cells. Oncogene. 2009;28:565–574. doi: 10.1038/onc.2008.404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fabian J, Lodrini M, Oehme I, Schier MC, Thole TM, Hielscher T, Kopp-Schneider A, Opitz L, Capper D, von Deimling A, et al. GRHL1 acts as tumor suppressor in neuroblastoma and is negatively regulated by MYCN and HDAC3. Cancer Res. 2014;74:2604–2616. doi: 10.1158/0008-5472.CAN-13-1904. [DOI] [PubMed] [Google Scholar]
- 5.Klein RH, Lin Z, Hopkin AS, Gordon W, Tsoi LC, Liang Y, Gudjonsson JE, Andersen B, Botchkarev V. GRHL3 binding and enhancers rearrange as epidermal keratinocytes transition between functional states. PLoS Genet. 2017;13:e1006745. doi: 10.1371/journal.pgen.1006745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Quan Y, Xu M, Cui P, Ye M, Zhuang B, Min Z. Grainyhead-like 2 promotes tumor growth and is associated with poor prognosis in colorectal cancer. J Cancer. 2015;6:342–350. doi: 10.7150/jca.10969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Quan Y, Jin R, Huang A, Zhao H, Feng B, Zang L, Zheng M. Downregulation of GRHL2 inhibits the proliferation of colorectal cancer cells by targeting ZEB1. Cancer Biol Ther. 2014;15:878–887. doi: 10.4161/cbt.28877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thiery JP, Acloque H, Huang RYJ, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 9.Nieto MA. Epithelial-Mesenchymal Transitions in development and disease: old views and new perspectives. Int J Dev Biol. 2009;53:1541–1547. doi: 10.1387/ijdb.072410mn. [DOI] [PubMed] [Google Scholar]
- 10.Lim J, Thiery JP. Epithelial-mesenchymal transitions: insights from development. Development. 2012;139:3471–3486. doi: 10.1242/dev.071209. [DOI] [PubMed] [Google Scholar]
- 11.Spaderna S, Schmalhofer O, Hlubek F, Jung A, Kirchner T, Brabletz T. Epithelial-mesenchymal and mesenchymal-epithelial transitions during cancer progression. Verh Dtsch Ges Pathol. 2007;91:21–28. [PubMed] [Google Scholar]
- 12.Schmalhofer O, Brabletz S, Brabletz T. E-cadherin, beta-catenin, and ZEB1 in malignant progression of cancer. Cancer Metastasis Rev. 2009;28:151–166. doi: 10.1007/s10555-008-9179-y. [DOI] [PubMed] [Google Scholar]
- 13.Zlobec I, Lugli A. Epithelial mesenchymal transition and tumor budding in aggressive colorectal cancer: tumor budding as oncotarget. Oncotarget. 2010;1:651–661. doi: 10.18632/oncotarget.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cieply B, Riley PA, Ivanov A, Pifer PM, Addison JB, Widmeyer J, Denvir J, Frisch SM. Abstract 3436: suppression of the epithelial-mesenchymal transition (EMT) by a wound-healing gene, grainyhead-like-2. Cancer Res. 2012;72:3436. doi: 10.1158/1538-7445.AM2012-3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Werner S, Frey S, Riethdorf S, Schulze C, Alawi M, Kling L, Vafaizadeh V, Sauter G, Terracciano L, Schumacher U, et al. Dual roles of the transcription factor grainyhead-like 2 (GRHL2) in breast cancer. J Biol Chem. 2013;288:22993–23008. doi: 10.1074/jbc.M113.456293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spaderna S, Schmalhofer O, Hlubek F, Berx G, Eger A, Merkel S, Jung A, Kirchner T, Brabletz T. A transient, EMT-linked loss of basement membranes indicates metastasis and poor survival in colorectal cancer. Gastroenterology. 2006;131:830–840. doi: 10.1053/j.gastro.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 17.Cieply B, Farris J, Denvir J, Ford H, Frisch SM. Epithelial-mesenchymal transition and tumor suppression are controlled by a reciprocal feedback loop between ZEB1 and grainyhead-like-2. Cancer Res. 2013;73:6299–6309. doi: 10.1158/0008-5472.CAN-12-4082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pieniążek M, Donizy P, Ziętek M, Szynglarewicz B, Matkowski R. The role of TGF-β-related signal transduction pathways in pathogenesis of epithelial-mesenchymal transition as a key element in cancer development and progression. Postepy Hig Med Dosw. 2012;66:583–591. doi: 10.5604/17322693.1009653. [DOI] [PubMed] [Google Scholar]
- 19.Morrison CD, Parvani JG, Schiemann WP. The relevance of the TGF-β Paradox to EMT-MET programs. Cancer Lett. 2013;341:30–40. doi: 10.1016/j.canlet.2013.02.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.


