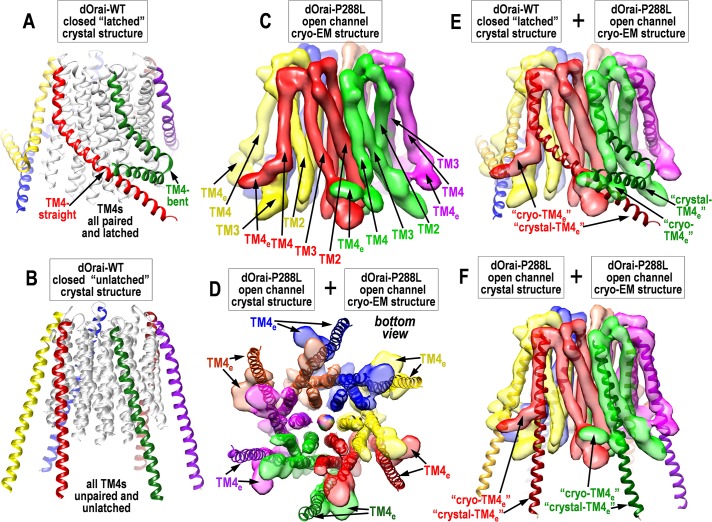
Fig 3. Details and comparison of crystal- and cryo-EM-derived dOrai structures.
(A) Side view of dOrai WT crystal structure of closed, “latched” channel, from the Zn2+-stabilized, paired-TM4e configuration [14]. TM4s (colored) have 3-fold symmetry because of antiparallel pairing of adjacent “straight” and “bent” TM4e configurations. (B) Side view of dOrai WT crystal structure of closed, “unlatched” channel from the “end-to-end”-stabilized configuration [11]. TM4s (colored) have 6-fold symmetry, are unpaired, straightened, and mostly detached from the channel core. (C) Side view of the cryo-EM structure of the dOrai-P288L open channel [12]. Note TM4e helices appear unpaired, bent in a clockwise direction, and closely associated with TM3s in adjacent subunits. Adjacent TM4s appear nonidentical, giving some outer 3-fold symmetry. (D) Comparison of bottom (cytosolic) view of crystal- and cryo-EM–derived structures of the dOrai-P288L open channel [12]. The crystal structure is from the end-to-end, unlatched configuration. Note there is close structural agreement for the inner core TM1, TM2, and TM3 helices, but unlatched TM4 helices are detached for the crystal structure and differ greatly, especially at the C-terminal TM4e helices that are critical STIM-binding sites. (E) Comparison of side views of the crystal-derived latched, closed channel (from panel A, but only TM4s shown) [14] and the cryo-EM–derived dOrai-P288L open channel (from panel C) [12]. Note the remarkable apparent difference in lower TM4 configuration—the straight anticlockwise-oriented TM4e in the crystal structure is bent and oriented clockwise in the cryo-EM structure. The bent TM4e in the crystal structure remains so in the cryo-EM structure but appears displaced clockwise. Both TM4es appear closely associated with TM3s in adjacent channel subunits. (F) Comparison of side views of the crystal-derived and cryo-EM-derived structures for the dOrai-P288L open channel (as in panel D) [12]. Only TM4s are shown for the crystal structure. Note the upper TM4s appear to be in close agreement, but the lower TM4s (including TM4es) are strikingly different—straight and unconnected for the crystal structure, and bent and associated with adjacent TM3s in the cryo-EM structure. cryo-EM, cryogenic electron microscopy; dOrai, Drosophila Orai; STIM, Stromal Interaction Molecule; TM, transmembrane; TM4e, TM4 extension; WT, wild type.