Figure 4: Factors influencing the distribution of m6A and m6Am on mammalian mRNAs.
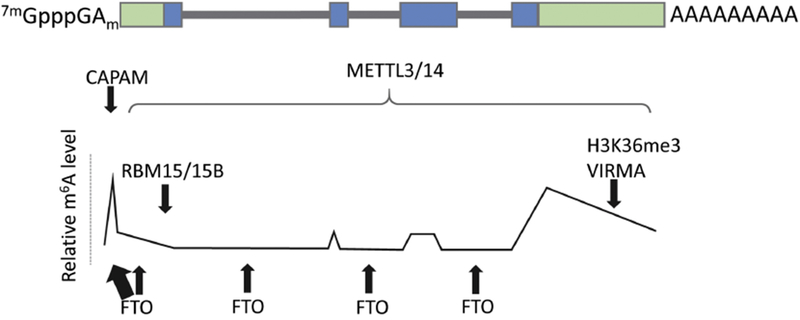
The overall m6A landscape in physiological conditions is determined primarily by the distribution of consensus motifs recognized by the methyltransferases (CAPAM, METTL3/14). However, it can be influenced by demethylases such as FTO, which prunes m6A from introns and across the transcript. Moreover, certain components of the MACOM, namely RBM15/15B and VIRMA, can favor methylation at the 5’ and 3’ ends respectively. Finally, there is evidence that histone modifications can influence m6A deposition.
