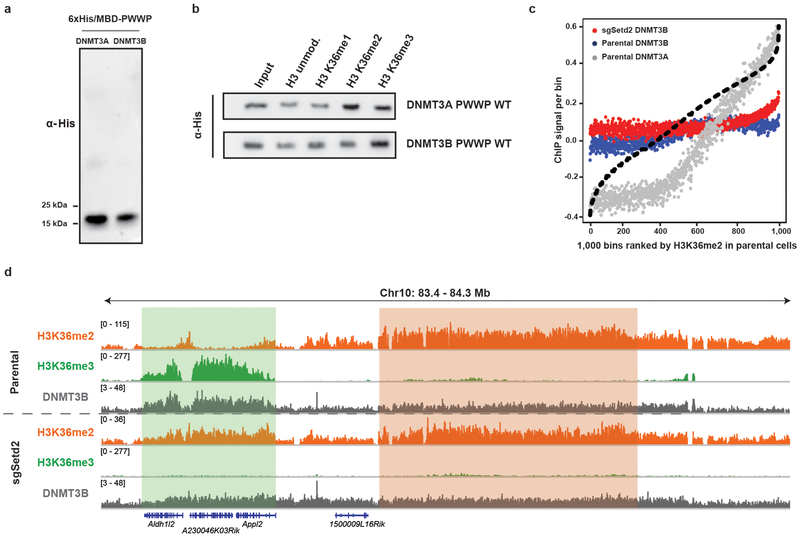
Extended Data Figure 8: Assessment of binding affinity between DNMT3B and H3K36me2 in vitro and in cells.
a) Immunoblots of His/MBP-tagged recombinant PWWP domains of DNMT3A and DNMT3B.
b) Immunoblots of recombinant His/MBP-tagged wild-type DNMT3A and DNMT3BPWWP domains bound to H3K36-modified recombinant nucleosomes following the in vitro pull-down assay.
c) ChIP-seq normalized reads per bin for DNMT3A1 (grey) and DNMT3B (blue) inparental mMSCs and DNMT3B (red) in sgSetd2 mMSCs relative to H3K36me2. To generate bins, 1kb genomic tiles were ranked by H3K36me2 enrichment in parental mMSCs and grouped into 1000 rank-ordered bins. Dashed line indicates H3K36me2 enrichment per bin.
d) Genome browser representation of ChIP-seq normalized reads for H3K36me2,H3K36me3, and DNMT3B in parental and sgSetd2 mMSCs at Chr10: 83.4-84.3 Mb, as indicated. Refseq genes are annotated at the bottom. Shaded areas indicate H3K36me2-enriched intergenic regions (orange) and H3K36me3-enriched genic regions (green) in parental cells. For H3K36me2 and H3K36me3, data are representative of two independent ChIP-seq experiments; for DNMT3B, ChIP-seq was performed once and an independent ChIP was performed in which genomic regions of selective enrichment and depletion were confirmed by qPCR.
The immunoblot data in a) and b) were independently repeated twice with similar results.
For gel source data, see Supplementary Fig. 1.