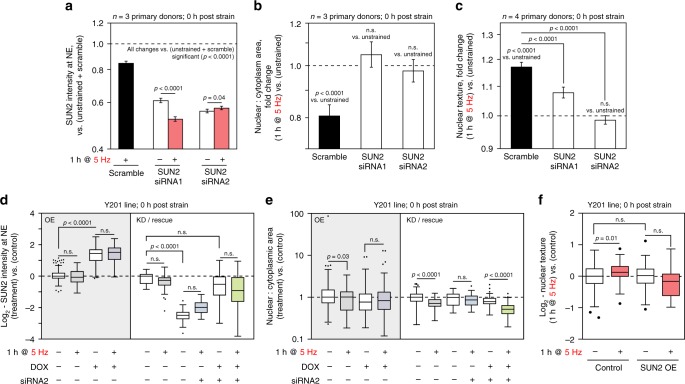
Fig. 8.
Responses to CTS are blocked by KD or OE of SUN2. a SUN2 levels quantified by IF at the NE in primary hMSCs, following SUN2 KD (comparing two siRNA sequences) and in combination with CTS (1 h, 2.6–6.2% strain at 5 Hz; n = 3 donors). Both siRNAs were effective against SUN2 (p < 0.0001). SUN2 level in the less potent KD (siRNA1) was further decreased by CTS (p < 0.0001); the more efficient KD (siRNA2) showed a small increase (p = 0.04). b Ratios of nuclear to cytoplasmic areas in primary hMSCs following SUN2 KD and CTS (n = 3 donors). SUN2 KD blocked the CTS-induced decrease to ratio (p < 0.0001 for scrambled vs. CTS). c Changes to nuclear texture in primary hMSCs following SUN2 KD and CTS (n = 4 donors). SUN2 KD significantly reduced strain-induced changes in nuclear texture (p < 0.0001 for SUN2 KD vs. scrambled and CTS). Plots show mean ± s.e.m.; p-values determined from linear modeling (ANOVA). See Supplementary Figs. 11a–f for data distributions and variation between donors. d Quantification of SUN2 at the NE in immortalised hMSCs with inducible SUN2 OE (p < 0.0001), SUN2 KD (siRNA2) (p < 0.0001) and induced rescue of SUN2 KD, immediately following 5 Hz CTS. e Ratios of nuclear to cytoplasmic areas in immortalised hMSCs with SUN2 OE, SUN2 KD (siRNA2) and SUN2 rescue, immediately following CTS. Ratios were significantly decreased in control cells following CTS (p = 0.03 and p < 0.0001), but not after OE or KD of SUN2. Rescue of SUN2 levels restored the response (p < 0.0001) (see Supplementary Figs. 11g–i). d and e were normalized to (−/−/−) controls; ≥35 cells analysed per condition. f Changes to nuclear texture in immortalised hMSCs following SUN2 OE and CTS (p = 0.01 for CTS vs. unstrained), ≥17 cells analysed per condition. Box-whisker plot center lines show medians, bounds of box show quartiles, whiskers show data spread and outliers determined by the Tukey method; significance determined by Dunnett’s multiple comparison tests. See Supplementary Table 1 for sample sizes