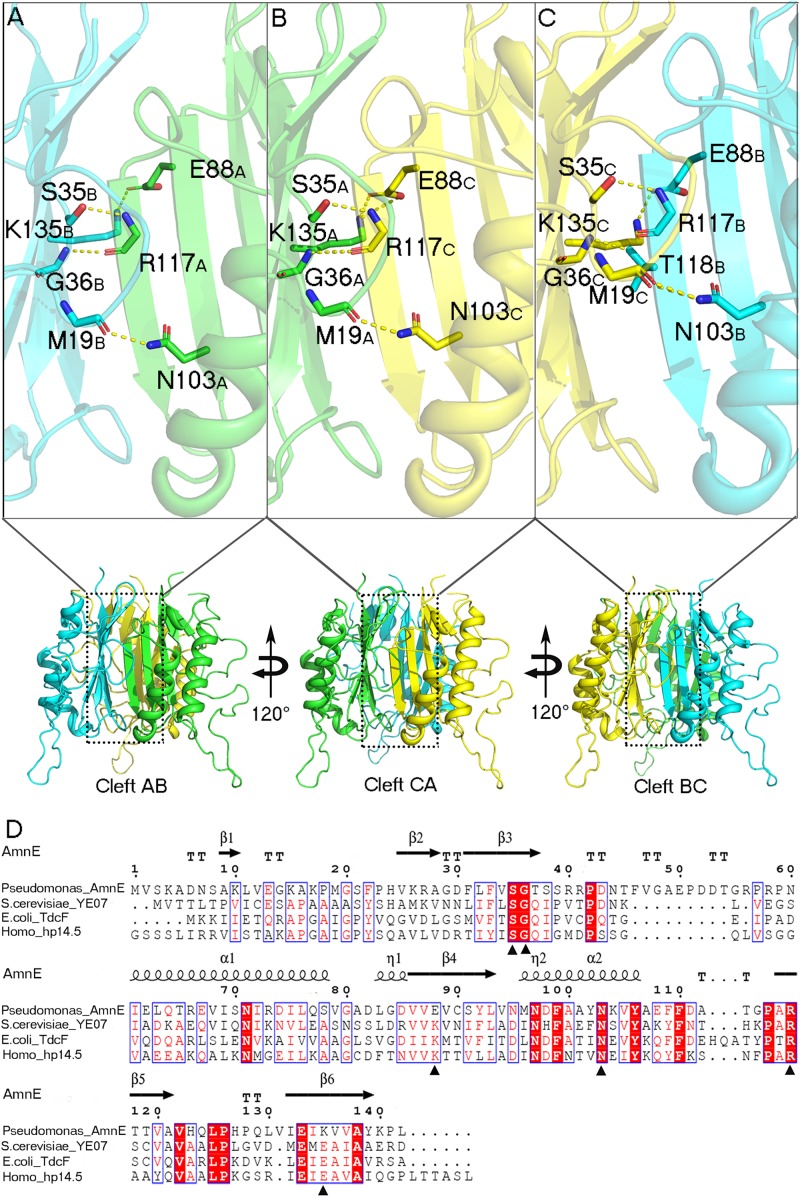
FIGURE 3.
Analyses of AmnE trimeric clefts. Conserved residues Ser35, Gly36, Glu88, Gln103, Arg117, and Lys135, are highlighted in the three clefts with sticks. (A) Interface of cleft AB. (B) Interface of cleft CA. (C) Interface of cleft BC. (D) Comparison of AmnE with the other three members of the YjgF/YER057c/UK114 family. Sequence alignment of AmnE from Pseudomonas sp. AP–3 with YE07 from Saccharomyces cerevisiae (34.00% sequence identity), TdcF from Escherichia coli (29.00% sequence identity), and hp14.5 from homo sapiens (28.72% sequence identity). Residues labeled with triangles are conserved residues involved in the ligand binding pocket. Sequences were aligned using the tool obtained from the web site (https://www.ebi.ac.uk/Tools/msa/clustalo/) and the alignment was presented using the online ESPript 3.0 server (http://espript.ibcp.fr/ESPript/ESPript/).