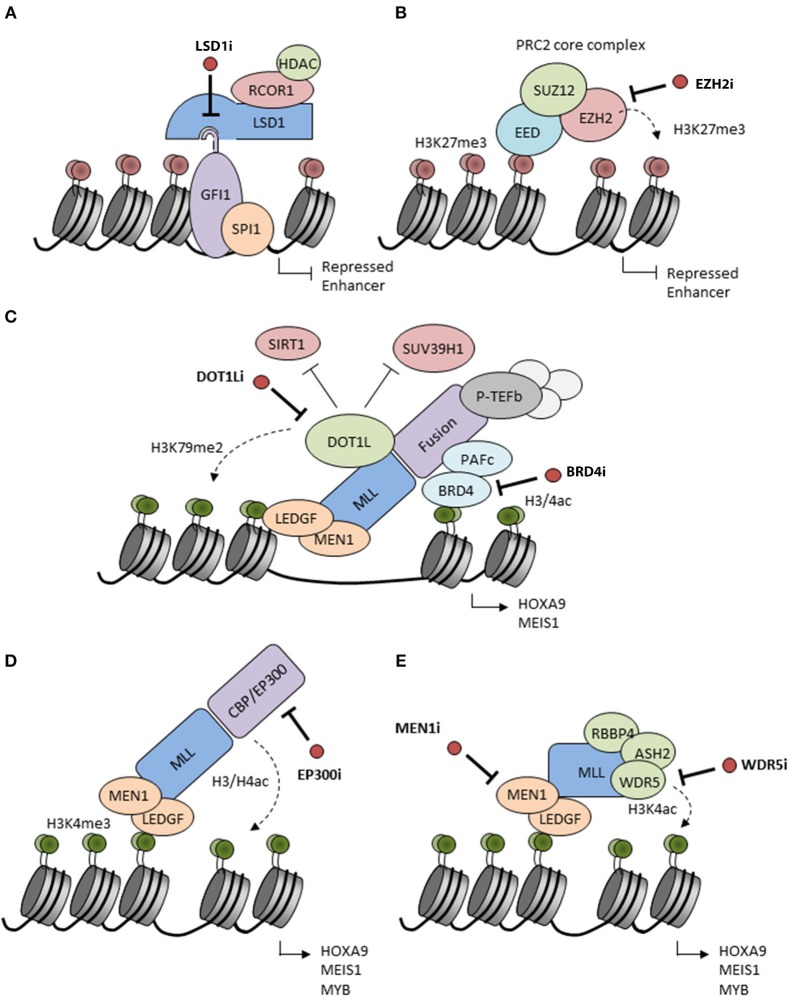
Figure 1.
Putative mechanisms of action of candidate epigenetic inhibitors. (A) LSD1 interacts with the SNAG domain of GFI1 recruiting repressors to chromatin. Inhibitors of LSD1 (LSD1i) disrupt the interaction and inactivate GFI1 leading to enhancer acetylation and activation. LSD1 inhibitors also inactivate the histone demethylase activity of LSD1 (not shown). (B) EZH2 catalyzes H3K27 methylation inducing transcriptional repression. This activity is blocked by S-adenosyl-methionine (SAM)-competitive inhibitors of EZH2 (EZH2i). (C) MLL fusion proteins form complexes on chromatin with Polymerase Associated Factor complex (PAFc) (which recruits Super Elongation Complex components), Positive Transcription Elongation Factor b (pTEFb) and other factors to facilitate the expression of MLL-driven target genes, such as HOXA9 and MEIS1. DOT1L is ectopically recruited by MLL fusions and adds activating H3K79me2 marks while reducing H3K9me2 repressive marks by inhibition of SUV39H1 and SIRT1. BRD4 recognizes H3K27ac marks and is essential for recruitment and stabilization of the MLL complex on chromatin. Inhibitors of the enzymatic activities of DOT1L (DOT1Li) or BRD4 (BRD4i) are considered to disrupt the MLL fusion protein complexes leading to the release of the differentiation block. (D) MLL may be fused to the histone acetyltransferases CBP or EP300 which are associated with H3/H4 acetylation and active gene transcription. CBP/EP300 bromodomain inhibition (EP300i) decreases H3K27 acetylation and chromatin accessibility at target promoters and enhancers. (E) The N-terminal part of the MLL complex associates with different proteins, such as LEDGF and Menin which stabilize the complex on chromatin. Proteins, such as RBBP5, ASH2L, and WDR5 interact with the MLL C-terminus to facilitate SET domain-mediated H3K4 methylation. Inhibition of these interactions (MEN1, WDR5i) disrupt the MLL complex and decrease expression of HOXA9 and MEIS1.