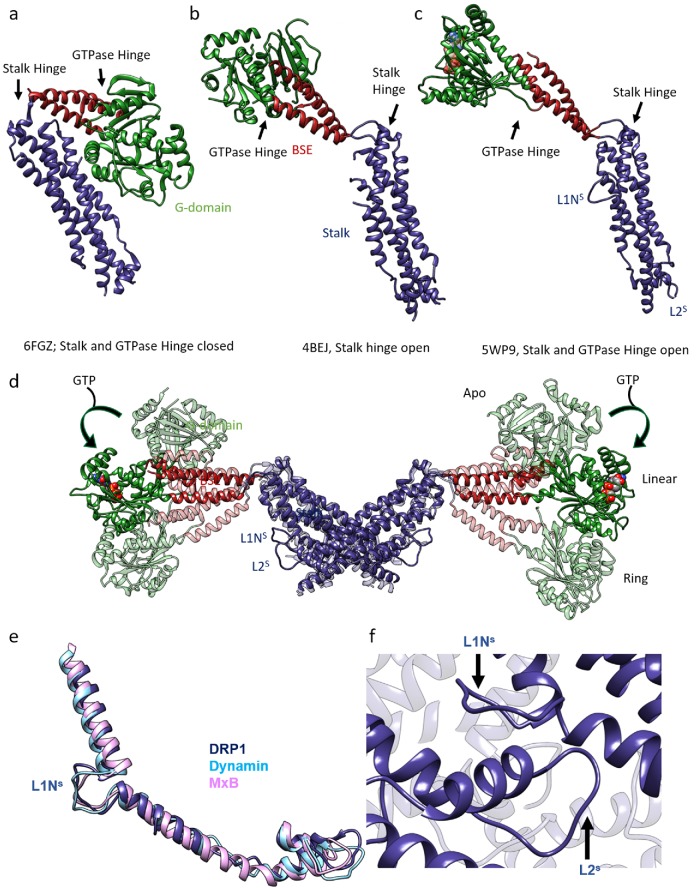
FIGURE 1:
Structural rearrangements within the DRP1 molecules upon nucleotide binding: (a) Cyanidioschyzon merolae Dnm1 crystal structure depicts a closed state of a DRP1 molecule. (b) Crystal structure of human DRP1 shows an open GTPase hinge from chain (A) where both the loops that constitute the GTPase hinge were ordered. (c) Cryo-EM structure of DRP1 determined with nucleotide and the MID49 receptor. The G-domain is rotated and loops L1NS and L2S are stabilized and visible. (d) Overlay of DRP1 dimers in the apo state (4BEJ, light shade, Fröhlich et al., 2013), the linear cryo-EM structure (5WP9, solid color), and the ring model (light shade, bent downward), as seen from Kalia et al. (2018), showing the range of movements exhibited by the G-BSE region relative to the stalk. The stalk is kept constant. (e, f) L1NS and L2S stabilization and interaction in dynamin family members: (e) Overlay of the region of the stalk that contains L1NS and L2S—from DRP1 cryo-EM structure (PDB ID: 5WP9), dynamin-3 crystal structure (PDB ID: 53AF), and MxB cryo-EM structure (PDB ID: 5UOT)—to depict the structural conservation of the region across dynamin family members. In each case, L1NS and L2S are stabilized by interdynamin and/or dynamin–receptor contacts. (f) L1NS and L2S mediate interdynamin contacts in the DRP1 cryo-EM structure (PDB ID: 5WP9).