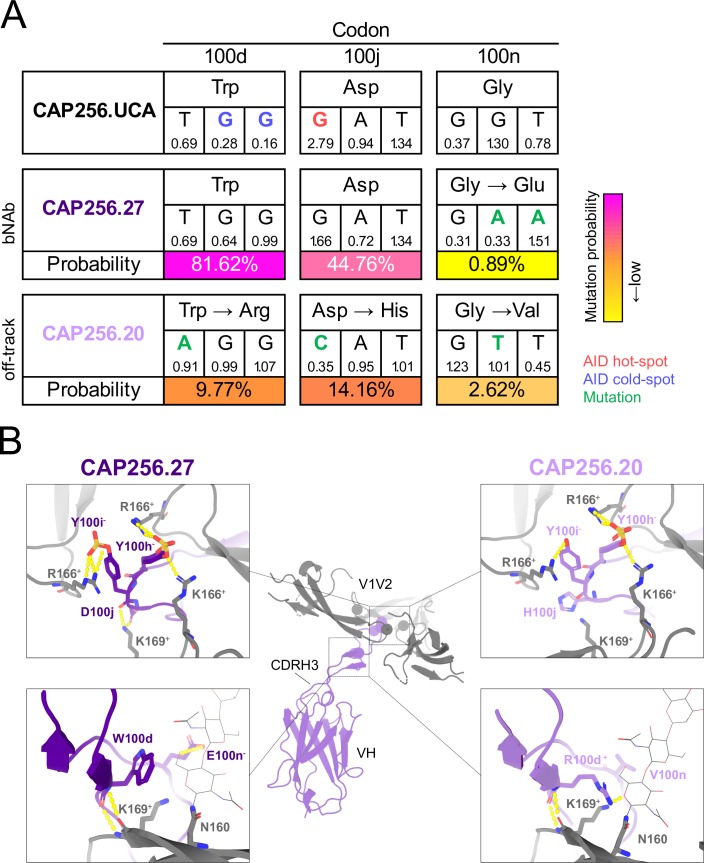
Fig 5. The probability and structural effect of the CAP256.20 off-track mutations W100dR, D100jH and G100nV.
(A) The germline (W100d, D100j and G100n) and mature nucleotide and amino acid sequences (CAP256.27 W100d, D100j and G100nE and CAP256.20 W100dR, D100jH and G100nV) at codons 100d, 100j and 100n are displayed. Blue and red text indicate AID cold- and hot-spots, respectively. Green text refers to mutations, and yellow to pink shading indicates low and high probability mutations, respectively. The mutability scores are below each nucleotide. (B) The sequence of CAP256.20 (light purple) and CAP256.27 (purple) were fitted to a structure of CAP256.25 and trimeric CAP256 34-week Env [26] in Swiss-PdbViewer (v4.1.0) and aligned and visualized in PyMOL (v2.0.2). The top panels shows the interaction between the CDRH3 YYD motif (purple/light purple) and the V2 epitope (grey). The bottom panel displays the CAP256.27 residues W100d and E100n (purple) and CAP256.20 R100d and V100n (light purple) in proximity to K169, + and—denotes positive and negative charge, respectively.