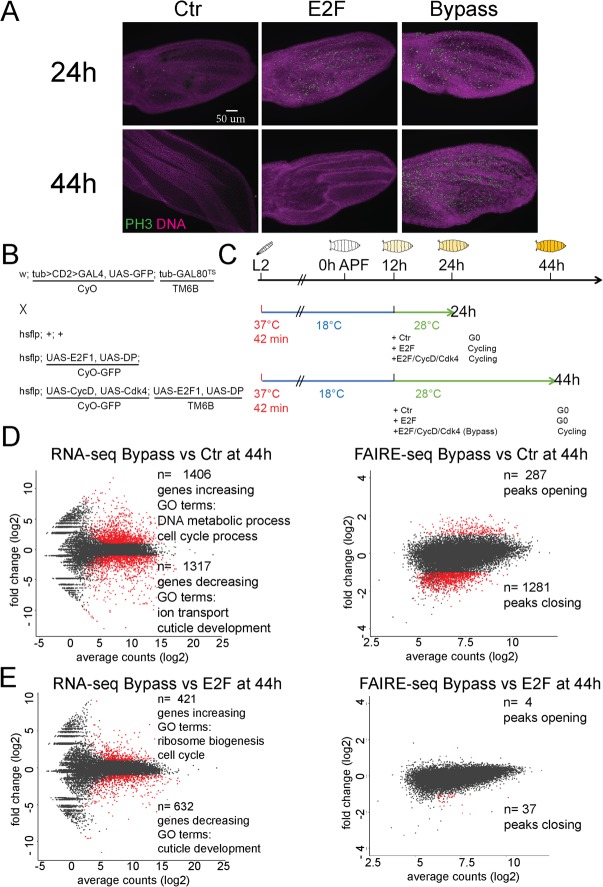
Fig 3. Global impacts of cell cycle exit disruption on gene expression and open chromatin.
(A) G0 can be delayed to 36 h or bypassed beyond 50 h through short-term expression of E2F or E2F + CycD/Cdk4. Transgenes were overexpressed in the dorsal layer of wing epithelia under the control of Apterous-Gal4/Gal80TS from 12 h APF. Twenty-four-hour and 44-h wings were immunostained for ph3. (B, C) Genotype and scheme of RNA-seq and FAIRE-seq experiments to disrupt cell cycle exit during metamorphosis. (D, E) MA plots of RNA and FAIRE changes comparing bypassed exit (E2F + CycD/cdk4) between Ctr (D) and delayed cell cycle exit (E2F) (E) at 44 h. Abundant changes in expression of cell cycle genes, ribosome biogenesis, and cuticle formation genes are observed, while chromatin accessibility is nearly identical between conditions in which cells enter a delayed G0 versus continue cycling. The underlying data in Fig 3D and 3E can be found within S3 Data. APF, after puparium formation; Cdk, cyclin-dependent kinase; Ctr, control; CycD, Cyclin D; CyO, Curly O balancer; DP, dimerization partner; E2F, E2F transcription factor; FAIRE, formaldehyde-assisted isolation of regulatory elements; FAIRE-seq, FAIRE sequencing; Gal80TS, temperature-sensitive Gal80; GO, Gene Ontology; hsflp, heatshock-flippase; MA plot, scatter plot onto M (log ratio) and A (mean average) scales; ph3, phosphohistone H3; RNA-seq, RNA sequencing; TM6B, TM6B balancer; UAS, upstream activation sequence.