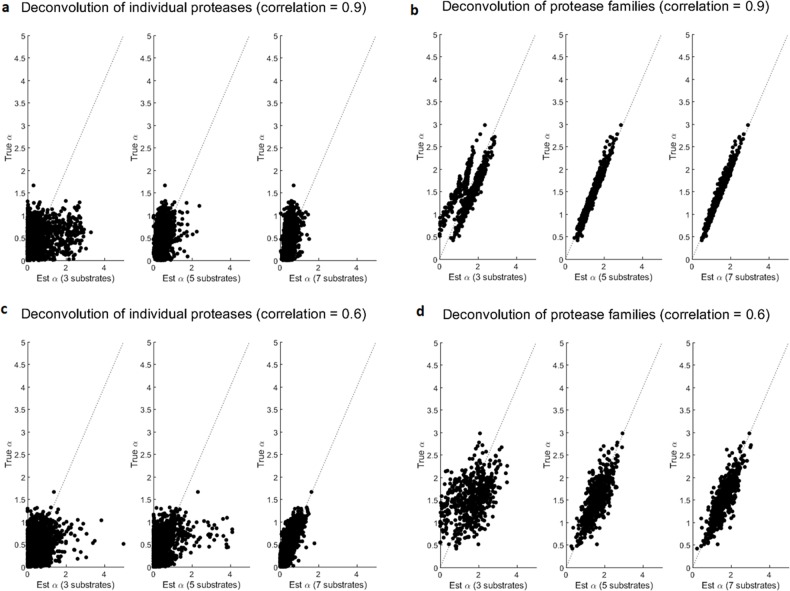
Fig 4. Comparison between deconvolution of individual proteases and deconvolution of protease families.
The x axis represented the estimated mixing coefficients. The y axis represented the true simulated mixing coefficients. Consider mixtures of 9 proteases from 3 independent families that contain highly correlated proteases within each family. (a) When deconvolving the 9 individual proteases, the estimated mixing coefficients for the individual proteases showed poor agreements with their true simulated values, regardless of whether 3, 5, or 7 substrates were used. (b) When deconvolving the 3 protease families, the estimated mixing coefficients for the protease families showed high agreement with the sum of the family members’ true mixing coefficients. (c-d) Simulation of protease families that contain moderately correlated proteases showed similar results, where deconvolution of individual proteases was difficult but deconvolution of protease families was accurate.