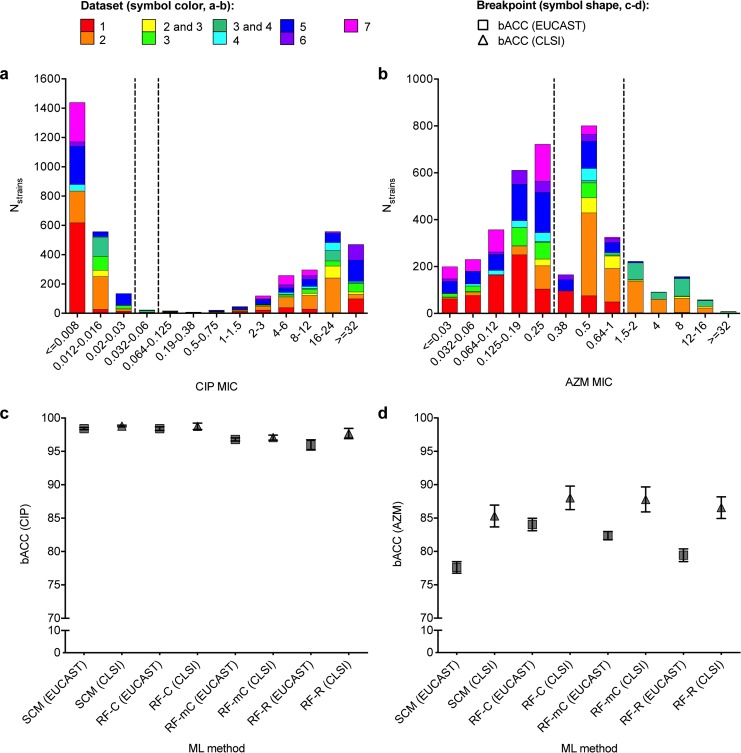
Fig 1. Differential performance of machine learning-based prediction models for ciprofloxacin and azithromycin resistance in gonococci.
Histograms showing the distributions of (a) ciprofloxacin (CIP) and (b) azithromycin (AZM) minimum inhibitory concentrations (MICs) in the gonococcal isolates assessed here. Bar color indicates the study or studies associated with the isolates. Dashed lines indicate the (a) EUCAST and CLSI breakpoints for non-susceptibility (NS, >0.03 μg/mL and >0.06 μg/mL, respectively) for CIP and the (b) EUCAST and CLSI breakpoints for non-susceptibility (>0.25 μg/mL and >1 μg/mL, respectively) for AZM. Note that there was some overlap in strains from the US between datasets 2 and 3 and in strains from Canada between datasets 3 and 4; such strains are indicated in (a) and (b) as belonging to datasets 2 and 3 and 3 and 4, respectively. Mean balanced accuracy (bACC) with 95% confidence intervals of predictive models for (c) CIP NS and (d) AZM NS trained and tested on the aggregate gonococcal dataset. Symbol colors in (a-b) indicate the datasets from which the training and testing sets were derived. Symbol shapes in (c-d) indicate the NS breakpoint. SCM, set covering machine; RF-C, random forest classification; RF-mC, random forest multi-class classification; RF-R, random forest regression.