New World (NW) arenaviruses are a diverse family of emerging zoonotic viruses that merit significant attention as important public health problems. The close genetic relationship of nonpathogenic NW arenaviruses with their highly pathogenic cousins suggests that few mutations may be sufficient to enhance virulence. The identification of molecular determinants of virulence of NW arenaviruses is therefore of great importance. Here we undertook a side-by-side comparison of the innate immune responses against the highly pathogenic Junin virus (JUNV) and the related nonpathogenic Tacaribe virus (TCRV) in human cells. We consistently found that TCRV induces a stronger type I interferon (IFN-I) response than JUNV. Transcriptome profiling revealed an overlapping pattern of IFN-induced gene expression and similar low sensitivities to IFN-I treatment. However, the double-stranded RNA (dsRNA)-dependent protein kinase R (PKR) contributed to the control of TCRV, but not JUNV, providing the first evidence for differential innate recognition and control of JUNV and TCRV.
KEYWORDS: arenavirus, innate immunity, interferon, protein kinase R, zoonosis
ABSTRACT
The New World (NW) arenaviruses are a diverse group of zoonotic viruses, including several causative agents of severe hemorrhagic fevers in humans. All known human-pathogenic NW arenaviruses belong to clade B, where they group into sublineages with phylogenetically closely related nonpathogenic viruses, e.g., the highly pathogenic Junin (JUNV) and Machupo viruses with the nonpathogenic Tacaribe virus (TCRV). Considering the close genetic relationship of nonpathogenic and pathogenic NW arenaviruses, the identification of molecular determinants of virulence is of great importance. The host cell’s innate antiviral defense represents a major barrier for zoonotic infection. Here, we performed a side-by-side comparison of the innate immune responses against JUNV and TCRV in human cells. Despite similar levels of viral replication, infection with TCRV consistently induced a stronger type I interferon (IFN-I) response than JUNV infection did. Transcriptome profiling revealed upregulation of a largely overlapping set of interferon-stimulated genes in cells infected with TCRV and JUNV. Both viruses were relatively insensitive to IFN-I treatment of human cells and induced similar levels of apoptosis in the presence or absence of an IFN-I response. However, in comparison to JUNV, TCRV induced stronger activation of the innate sensor double-strand RNA-dependent protein kinase R (PKR), resulting in phosphorylation of eukaryotic translation initiation factor eIF2α. Confocal microscopy studies revealed similar subcellular colocalizations of the JUNV and TCRV viral replication-transcription complexes with PKR. However, deletion of PKR by CRISPR/Cas9 hardly affected JUNV but promoted TCRV multiplication, providing the first evidence for differential innate recognition and control of pathogenic and nonpathogenic NW arenaviruses by PKR.
IMPORTANCE New World (NW) arenaviruses are a diverse family of emerging zoonotic viruses that merit significant attention as important public health problems. The close genetic relationship of nonpathogenic NW arenaviruses with their highly pathogenic cousins suggests that few mutations may be sufficient to enhance virulence. The identification of molecular determinants of virulence of NW arenaviruses is therefore of great importance. Here we undertook a side-by-side comparison of the innate immune responses against the highly pathogenic Junin virus (JUNV) and the related nonpathogenic Tacaribe virus (TCRV) in human cells. We consistently found that TCRV induces a stronger type I interferon (IFN-I) response than JUNV. Transcriptome profiling revealed an overlapping pattern of IFN-induced gene expression and similar low sensitivities to IFN-I treatment. However, the double-stranded RNA (dsRNA)-dependent protein kinase R (PKR) contributed to the control of TCRV, but not JUNV, providing the first evidence for differential innate recognition and control of JUNV and TCRV.
INTRODUCTION
Mammalian arenaviruses are a diverse family of emerging viruses that includes several causative agents of severe viral hemorrhagic fevers with high mortality in humans (1, 2). Based on their antigenic properties, phylogeny, and geographic distribution, mammalian arenaviruses are divided into Old World (OW) and New World (NW) groups (3). The OW arenaviruses include the prototypic lymphocytic choriomeningitis virus (LCMV) and the highly pathogenic Lassa virus (LASV) endemic in Western Africa. The phylogenetically more diverse NW arenaviruses are subdivided into clades A, B, and C, found in South America, and clade D, which is restricted to the Northern American continent (4). All known human-pathogenic NW arenaviruses belong to clade B and include Junin (JUNV), Machupo (MACV), Guanarito (GTOV), Sabia (SABV), and Chapare (CHAV) virus. Arenaviruses of clades A and C have so far not been associated with human pathology, whereas the disease potential of clade D viruses remains unclear.
Mammalian arenaviruses are enveloped negative-strand RNA viruses with a nonlytic life cycle confined to the cytosol (5). The mammarenaviral genome is composed of two RNA segments, a small (S) RNA that encodes the envelope glycoprotein precursor (GPC) and the nucleoprotein (NP) and the L segment coding for the matrix protein (Z) and the viral polymerase (L). The arenavirus GPC undergoes proteolytic processing to yield GP1, involved in receptor binding, and GP2, which mediates viral fusion. The principal hosts for NW arenaviruses are rodents of the subfamilies Neotominae and Sigmodontinae of the Cricetidae family, with the exception of TCRV, which seems to be carried by fruit bats (4). In their natural reservoir hosts, NW arenaviruses are maintained by persistent infection and vertical transmission from infected mothers to offspring in utero, resulting in a carrier state (1). Human infection with pathogenic NW arenaviruses occurs mainly via reservoir-to-human transmission, involving inhalation of aerosolized contaminated rodent excreta or direct contact with infected rodents (6). Due to their transmissibility via aerosol and high lethality, hemorrhagic NW arenaviruses are considered category A select agents by the Centers of Disease Control and Prevention (7).
Pathogenic NW arenaviruses do not cluster within a defined sublineage of clade B but group with phylogenetically closely related nonpathogenic viruses. The highly pathogenic JUNV and MACV, for example, group with the nonpathogenic TCRV, which causes only mild febrile illness in humans (4). This close genetic relationship of nonpathogenic NW arenaviruses with their highly pathogenic cousins suggests that a relatively low number of adaptive mutations may be sufficient to enhance virulence in humans. The identification of the molecular determinants underlying the distinct virulence of pathogenic and nonpathogenic NW arenaviruses is therefore of great importance for epidemiology and public health.
A first major barrier for zoonotic viruses is the recognition of a suitable receptor(s) at the surface of susceptible human cells. The clade B NW arenaviruses evolved to hijack the conserved cargo-receptor transferrin receptor 1 (TfR1) for host cell entry, and human disease potential strongly correlates with their ability to recognize the human receptor form (8–11). In the absence of a functional TfR1 orthologue, NW arenaviruses can use evolutionary conserved phosphatidylserine (PS) receptors of the Tyro3/Axl/Mer (TAM) and T-cell immunoglobulin and mucin domain-containing (TIM) protein family for cell entry via “apoptotic mimicry” (12, 13). Once zoonotic NW arenaviruses have penetrated into the cytosol, they encounter the host cell’s powerful innate antiviral immune defense that represents another crucial barrier. The major pathogen recognition receptors implicated in innate detection of arenaviruses in the cytosol are the ubiquitously expressed RNA helicases retinoic acid-inducible gene I (RIG-I) and melanoma differentiation-associated gene 5 (MDA5) (14–18). RIG-I detects single-stranded RNA with 5′ triphosphates and short stretches of double-stranded RNA (dsRNA) as a “danger signal,” whereas MDA5 is activated by longer dsRNA (19). Upon activation, RIG-I and MDA5 induce downstream signaling via the mitochondrial adaptor of antiviral signaling (MAVS) (20–23). Activation of MAVS results in assembly of a signaling complex that includes the classical IκB kinases IKKα/IKKβ/NEMO involved in activation of nuclear factor κB (NF-κB). Recruitment of TANK-binding kinase (TBK1) and IKKε activates interferon regulatory factor 3 (IRF3) and IRF7. Simultaneous activation of IRF3/7 and NF-κB induces the expression and secretion of type I interferons (IFN-I) that regulate transcription of hundreds of interferon-stimulated genes (ISGs), establishing an antiviral state. In addition, IFN-I stimulate and regulate cells of innate and adaptive immunity, including natural killer cells, macrophages, dendritic cells, and T cells.
The OW arenaviruses LCMV and LASV can efficiently evade innate immunity in many cell types (24–27), whereas infection with the NW viruses JUNV, MACV, and TCRV induces significant IFN-I levels (18, 26–29). The NPs of arenaviruses act as major IFN-I antagonists by preventing activation of IRF3 and NF-κB (24, 25, 30–36). The IFN-I-suppressive activity of arenavirus NP is linked to a 3′–5′ exoribonuclease activity located within its C-terminal region, which likely degrades immunogenic viral RNA species, removing the “danger signal” (30–36). The structural motifs required for the 3′–5′ exoribonuclease activity are highly conserved among arenaviruses and likely crucial for viral persistence in reservoirs (31–34, 36). However, early comparative studies using recombinant NPs of different arenaviruses revealed stronger IFN-I suppression and NF-κB inhibition by NPs of OW arenaviruses and the pathogenic JUNV and MACV than by the NP of the nonpathogenic TCRV (25, 28). Moreover, the Z proteins of human-pathogenic arenaviruses bind RIG-I and interfere with MAVS activation, whereas Z proteins from nonpathogenic arenaviruses like TCRV cannot (37, 38).
Another important innate sensor for RNA viruses is the dsRNA-dependent protein kinase R (PKR). Recent studies demonstrated that the NW arenaviruses JUNV and MACV, but not the OW arenavirus LASV, activate PKR (26, 39). In JUNV-infected cells, PKR colocalizes with the viral replication-transcription complex in the cytosol (39, 40). Interestingly, depletion of PKR reduces the production of infectious JUNV and MACV particles (26). Early activation of PKR by JUNV and MACV restricts translation of key ISGs, allowing optimal viral multiplication, indicating that these pathogenic viruses evolved to hijack PKR for their own needs. The specific role of PKR in infection with nonpathogenic NW arenaviruses is, however, unknown.
In summary, several lines of evidence suggest important differences between the innate responses against pathogenic and nonpathogenic NW arenaviruses, which likely contribute to their distinct disease potentials. To confirm and extend these studies, we undertook a side-by-side comparison of the innate immune responses against JUNV and TCRV in human cells. We consistently found that TCRV induces a stronger IFN-I response than JUNV. Despite these quantitative differences, transcriptome profiling revealed a largely overlapping pattern of IFN-induced gene expression after infection with JUNV and TCRV. The two viruses showed similar low sensitivities to IFN-I treatment. However, JUNV and TCRV differentially activated PKR, which contributed to control of TCRV, but not JUNV, providing the first evidence for distinct recognition by this important innate sensor.
RESULTS
TCRV infection induces a stronger IFN-I immune response than JUNV infection.
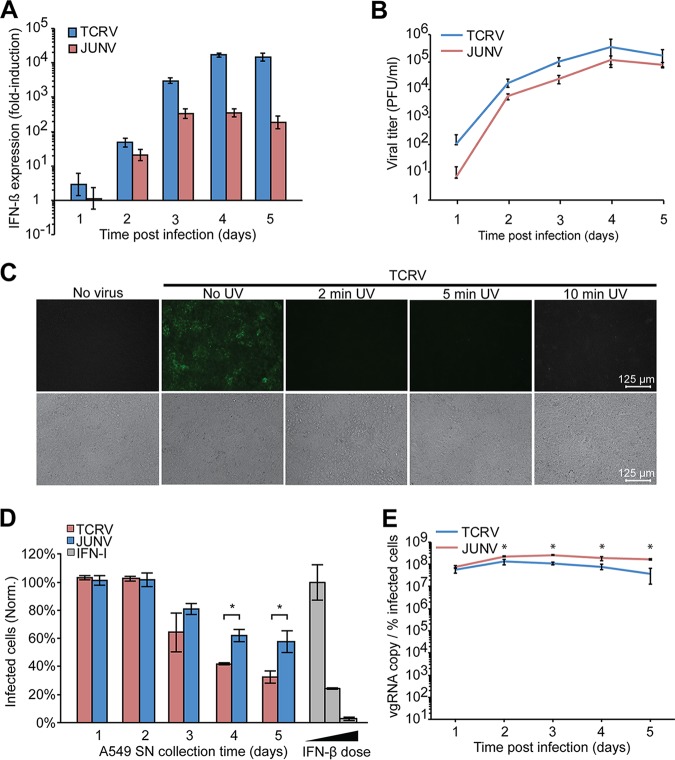
For our comparative study of innate immunity against pathogenic and nonpathogenic NW arenaviruses, we chose JUNV, which represents the most important pathogen among NW arenaviruses, and the phylogenetically closely related TCRV, a classical experimental model for nonpathogenic NW arenaviruses (4). Since work with live JUNV requires biosafety level 4 (BSL4) containment, we employed the JUNV vaccine strain Candid1, which can be manipulated under BSL2 (41). Reverse genetics mapped the attenuation of JUNV Candid1 exclusively to the GPC, with little contributions by NP, Z, or L (42, 43). Mutations in Candid1 NP lie outside regions critical for its 3′–5′ exoribonuclease function (31, 44), and the Z sequence remained unchanged in comparison to the pathogenic parental JUNV XJ13 (44). Accordingly, infection of human cells with the pathogenic JUNV isolate Romero and JUNV Candid1 results in similar IFN-I responses and ISG expression (18), making JUNV Candid1 a suitable BSL2 surrogate to study innate immune responses. To avoid artifacts caused by the passage history of viruses, we plaque purified JUNV Candid1 and TCRV strain 11573, followed by amplification of clonal isolates in IFN-I-deficient BHK21 cells. Viruses were precipitated by polyethylene glycol, followed by purification through a sucrose cushion by ultracentrifugation. Since zoonotic transmission of NW arenaviruses can occur via aerosols (6), we chose the human lung epithelial cell line A549, which has been widely used to study arenavirus innate immunity (18, 24–28, 39, 40, 45), as a cell culture model. A549 cells express human TfR1 in combination with the TAM receptor Axl and TIM-1 (46, 47), allowing entry of TCRV via the PS receptors Axl and TIM-1 in the absence of a functional TfR1 orthologue (12). First, we infected monolayers of A549 cells with plaque-purified JUNV Candid1 and TCRV at a low multiplicity of infection (MOI, 0.1) and monitored IFN-β mRNA levels using real-time quantitative PCR (RT-qPCR). Consistent with previous studies, both JUNV Candid1 and TCRV induced robust transcription of IFN-β in A549 cells 2 days postinfection (18, 28). Over time, we consistently observed higher IFN-β mRNA levels in A549 cells infected with TCRV than in those infected with JUNV Candid1 (Fig. 1A). Detection of infectious virus in conditioned cell culture supernatants by an immunofocus assay (IFA) revealed slightly higher titers for TCRV than for JUNV Candid1 (Fig. 1B).
FIG 1.
TCRV infection triggers a stronger IFN-I immune response than JUNV Candid1 infection. (A) Detection of IFN-β mRNA levels in A549 cells infected with JUNV and TCRV. Monolayers of A549 cells were infected with TCRV and JUNV Candid1 at an MOI of 0.1 PFU/cell or were mock infected. At the indicated time points, cells were harvested, total RNA was extracted, and mRNA of IFN-β and GAPDH was quantified by RT-qPCR. Data shown are fold induction above background relative to that of mock-infected cells. Data are means ± SD (n = 3). (B) Supernatants from cells described for panel A were tested for viral titer by IFA, and data are means ± SD (n = 3). (C) Inactivation of virus by UV irradiation. TCRV (106 PFU) was exposed to UV irradiation for the indicated times and used to infect fresh VeroE6 monolayers. After 48 h, cells were fixed and stained for TCRV NP by IFA. Images are representative of three biological replicates. Bar, 125 μm. (D) Detection of IFN-I activity by bioassay. Conditioned supernatants collected (days 1 to 5) from infected A549 cells were UV inactivated for 2 min and used to pretreat VeroE6 cells for 16 h. As a positive control, recombinant IFN-I (1,000, 100, and 10 IU/ml; diluted in supernatant from uninfected cells) was subjected to the same UV exposure and used in parallel with tested samples. Pretreated VeroE6 cells were infected with VSV for 8 h, fixed, and stained for VSV M protein by IFA. Results are means ± SD (n = 3) of results from one representative experiment out of two independent experiments (P < 0.001). Norm, normalized. (E) Detection of viral genomic RNA (vgRNA) in infected cells. Total RNA samples from panel A were subjected to RT-qPCR using specific probe/primers sets for JUNV Candid1 and TCRV genomic S RNA detection. Cells from parallel specimens in panel A were examined for NP expression by flow cytometry. The ratios of viral RNA copies to percentage of infected cells were determined as detailed in Materials and Methods. Statistical t tests were performed with Excel software; *, P < 0.001. Data are means ± SD (n = 3) of the results of one out of three independent experiments.
Previous studies demonstrated that conditioned cell culture supernatants from TCRV-infected A549 cells contained IFN-I levels capable of inducing an antiviral stage in IFN-I-deficient Vero cells (28). To validate efficient UV inactivation of TCRV in our system, we subjected 106 PFU of TCRV to UV treatment for the indicated times to inactivate the virus (Fig. 1C). The UV-treated samples were added to fresh VeroE6 monolayers, and productive infection was detected after 48 h by IFA using a monoclonal antibody (MAb) to TCRV NP (Fig. 1C). To detect bioactive IFN-I-conditioned cell culture supernatants, we exposed supernatants from TCRV- or JUNV-infected A549 cells to UV irradiation for 2 min and used them to treat VeroE6 cells that are unable to produce IFN-I but remain capable of mounting an antiviral state in response to exogenously added IFN-I (48). For a positive control, we included recombinant human IFN-I. After 16 h of pretreatment, VeroE6 monolayers were infected with vesicular stomatitis virus (VSV), which is highly sensitive to IFN-I. Monolayers were fixed after 8 h, and productive VSV infection was detected by IFA using a MAb to the VSV matrix (M) protein. Consistent with previous reports (28), VeroE6 cells treated with conditioned cell culture supernatants from TCRV-infected A549 cells restricted replication of VSV (Fig. 1D), confirming the production of bioactive IFN-I by TCRV-infected A549 cells.
The observed higher TCRV titers suggested more extensive viral replication at the cellular level, which may explain the stronger IFN-I induction (Fig. 1A). To address this possibility, we quantified the intracellular levels of viral genomic RNA, which represents a major immunogenic viral RNA species (49), by RT-qPCR and normalized the result to the percentage of infected cells as assessed by detection of viral NP via flow cytometry (for details, see Materials and Methods). Rather unexpectedly, we observed higher intracellular levels of viral genomic RNA for JUNV Candid1 than for TCRV after 24 h of infection (Fig. 1C). This suggested that the observed quantitative differences in the IFN-I responses to JUNV Candid1 and TCRV were not simply a consequence of different replication levels.
JUNV Candid1 blocks innate signaling more efficiently than TCRV.
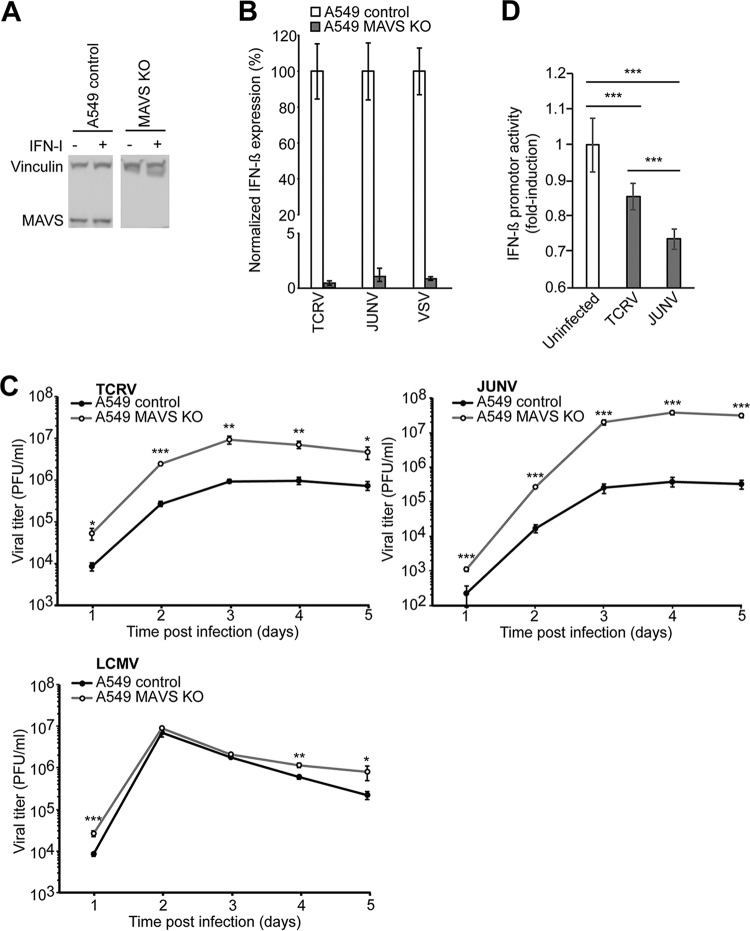
Previous studies have implicated the RIG-I/MAVS pathway in innate sensing of JUNV (18), whereas the innate sensors for TCRV have remained unknown. To address the role of RIG-I/MAVS in innate sensing of TCRV, we deleted MAVS from A549 cells using CRISPR/Cas9 technology as detailed in Materials and Methods and verified MAVS depletion by Western blot analysis (Fig. 2A). Monolayers of MAVS knockout (KO) and control A549 cells were infected with JUNV Candid1 and TCRV, and IFN-β mRNA was detected after 4 days by RT-qPCR. As a benchmark, we included VSV, which induces a robust MAVS-dependent IFN-I response (50). Deletion of MAVS almost completely abrogated IFN-β induction in cells infected with JUNV Candid1, TCRV, and VSV, confirming a key role of MAVS-dependent pathways in their innate detection (Fig. 2B). Deletion of MAVS increased JUNV Candid1 and TCRV multiplication, confirming a role of MAVS in their innate control (Fig. 2C). As expected, MAVS deficiency only mildly affected the OW arenavirus LCMV, which efficiently suppresses innate immune signaling (24, 28, 31) (Fig. 2C).
FIG 2.
Detection of JUNV Candid1 and TCRV by MAVS. (A) MAVS expression in A549 control and A549 MAVS KO cells. Cells were incubated with or without 10,000 IFN IU/ml and lysed after 24 h. Total proteins were separated by SDS-PAGE and probed with anti-MAVS MAb and anti-vinculin MAb by Western blotting. (B) IFN-I induction in A549 control and A549 MAVS KO cells after infections with TCRV, JUNV Candid1, and VSV. Both cell lines were infected with the indicated viruses at an MOI of 0.1 PFU/cell, and IFN-β mRNA levels were quantified by RT-qPCR 4 days postinfection (TCRV and JUNV Candid1) or 20 h postinfection (VSV). All values were normalized to the value for A549 control cells. Data are means ± SD (n = 3) of results from one representative example out of two independent experiments. (C) Growth kinetics of LCMV, TCRV, and JUNV Candid1 in A549 control and A549 MAVS KO cells. Cells were infected with TCRV, JUNV Candid1, or LCMV at an MOI of 0.1 PFU/cell, and viral progeny production was assessed over 5 days by IFA. Data are means ± SD (n = 3), with one representative example of three independent experiments shown. Statistical significance (t test) is indicated as follows: *, P < 0.01; **, P < 0.001; ***, P < 0.0001. (D) Differential IFN-I suppression by JUNV Candid1 and TCRV. HEK293T cells were infected with TCRV or JUNV Candid1 at an MOI of 0.1 PFU/cell or were mock infected. After 24 h, cells were transfected with the plasmid pIRES-LUC expressing a luciferase reporter under the control of an IFN-β promoter and assayed for luminescence activity after another 24 h. Data are means ± SD (n = 3) of the results of three independent experiments. Statistical t tests were performed, and significance is indicated as follows: *, P < 0.01; **, P < 0.001; ***, P < 0.0001.
The higher IFN-I levels in TCRV infection than in JUNV Candid1 infection, despite comparable intracellular levels of viral RNA (Fig. 1A and C), was in line with the previously demonstrated stronger IFN-I suppression by recombinant JUNV NP and Z (25, 28, 38). We next sought to confirm this in the context of productive arenavirus infection. Previous studies on innate immune suppression by arenavirus infection frequently used superinfection with RNA viruses like VSV or Sendai virus (24, 25, 28). However, this classical approach was complicated by the significant IFN-I levels induced by JUNV and TCRV in our system (Fig. 1). As an alternative innate “danger signal,” we therefore used transfection of cytosolic dsDNA of bacterial origin, which can serve as a template for Pol-III-driven synthesis of dsRNA species capable of activating RIG-I/MAVS, as well as the cGAS/STING pathway (51–53). Briefly, we infected monolayers of HEK293T cells with TCRV or JUNV Candid1 or mock infected them for 24 h, resulting in >95% of infected cells assessed by flow cytometry (data not shown). We subsequently transfected cells with a reporter plasmid of bacterial origin expressing firefly luciferase under the control of an IFN-β promoter. Detection of luciferase activity by a luminescence assay after 24 h revealed robust induction of IFN-β promoter activity in mock-infected cells triggered by plasmid DNA transfection, as expected (Fig. 2B). Previous infection with JUNV Candid1 reduced IFN-β promoter activity 2-fold, indicating active immunosuppression (Fig. 2D). In contrast, the presence of TCRV resulted in a less-pronounced reduction of IFN-β promoter activity (Fig. 2D). In summary, the data confirm differential inhibition of innate signaling during JUNV Candid1 and TCRV infections, consistent with the previously reported differences in IFN-I antagonist function of recombinant JUNV and TCRV NP and Z (28, 38).
Infection with JUNV Candid1 and TCRV induce overlapping sets of ISGs.
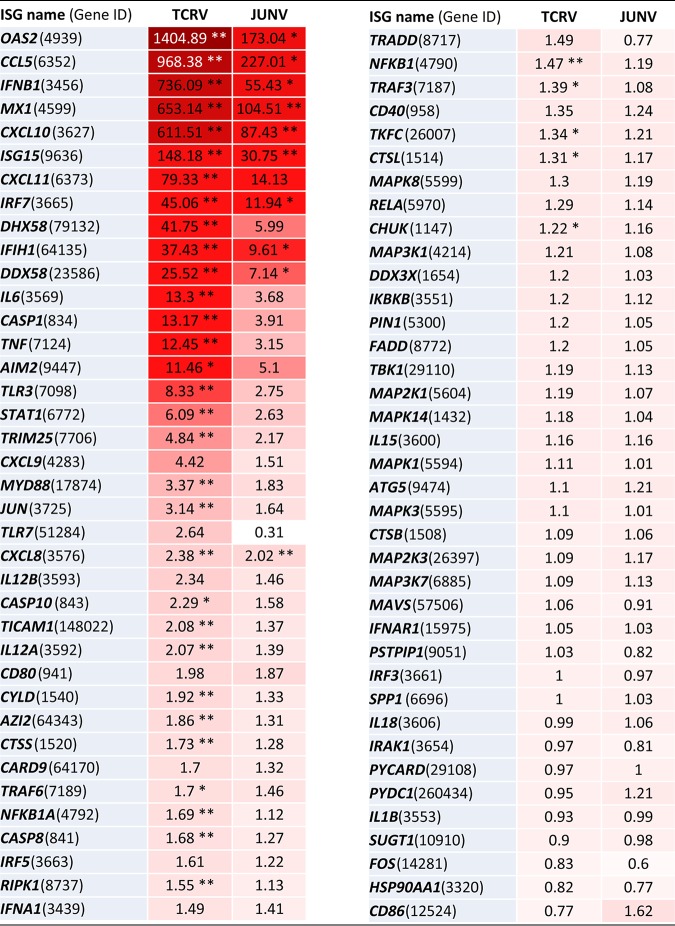
We then performed a more detailed analysis of the IFN-I-induced ISG expression pattern in response to infection with JUNV Candid1 and TCRV. To this end, we infected monolayers of A549 cells with JUNV Candid1 and TCRV and extracted the total RNA after 48 h, a time point when both viruses induce robust IFN-β levels (Fig. 1A). We then quantified the expression levels of 76 selected ISGs using the human antiviral response RT2 Profiler PCR array, as detailed in Materials and Methods. Examination of the ISG expression profiles for JUNV Candid1 and TCRV infection confirmed the previously observed quantitative differences in IFN-β induction (Table 1). Among the 76 ISGs covered, 10 genes showed significant upregulation in JUNV Candid1 infection, whereas TCRV infection significantly affected expression of 36 ISGs (Table 1). We consistently observed upregulation of the RIG-I transcript DDX58 (25.52-fold) and the MDA5 transcript IFIH1 (37.43-fold) in TCRV-infected cells, whereas JUNV Candid1 infection only mildly increased their transcription levels (7.14- and 9.61-fold, respectively) (Table 1). TCRV but not JUNV infection induced expression of the laboratory of genetics and physiology 2 (LGP2) transcript DHX58 (41.75-fold). The RIG-I helicase variant LGP2 lacks the N-terminal caspase recruitment domains (CARD) responsible for interaction with MAVS. Initial studies implicated LGP2 in the negative regulation of RIG-I/MAVS-mediated signaling (54). However, studies in knockout mice revealed positive regulatory functions of LGP2 on innate responses to several RNA viruses (55–57). LGP2 is not essential for induction of innate immune defenses to LCMV in mice but seems required for an optimal antiviral CD8 T cell response (57). The role of LGP2 in NW arenavirus infection is currently unknown. In contrast to the findings for the upstream RIG-I helicases, we did not detect upregulation of MAVS or any of its downstream signaling molecules included in the array. Among transcription factors involved in expression of IFN-I and cytokines, we observed induction only of the IFN-I-regulated IRF7 in TCRV infection (45.06-fold) and JUNV Candid1 infection (11.94-fold) (Table 1), whereas expression of the latent transcription factors IRF3 and NF-κB were not altered. Despite significant differences in the amplitude of the signals, statistical analysis of the relative expression levels of analyzed ISGs revealed a similar overall pattern between TCRV and JUNV infections (Table 1). Similar to previous studies with the pathogenic JUNV Romero and MACV (26), infection of A549 cells with TCRV and JUNV Candid1 significantly increased expression of the 2′–5′ oligoadenylate synthetase 2 gene (OAS-2), MX1, and ISG15, which critically contribute to the antiviral state (Table 1). We further observed upregulation of the transcription of cytokine genes CCL5, CXCL10, and CXCL11. In summary, an overlapping set of ISGs was upregulated in response to TCRV and JUNV Candid1 in our system. The data suggest that the viral mechanisms capable of modulating the innate immune response may be similar for the two viruses but seem to operate more efficiently in JUNV.
TABLE 1.
ISG profile upon TCRV and JUNV infectiona
A549 cells were infected with TCRV and JUNV at an MOI of 0.1 PFU/cell. After 48 h, total cellular RNA was extracted and transcriptome profiling was performed using the human antiviral response RT2 Profiler PCR array (PAHS-122ZG; Qiagen) as detailed in Materials and Methods. Values represent fold induction ± SD (n = 4) relative to that of mock-infected controls. The intensity of the red background color is proportional to fold induction. *, P < 0.01; **, P < 0.001 (t test between infected samples and mock-infected controls).
JUNV Candid1 and TCRV show similar low sensitivities to IFN-I treatment.
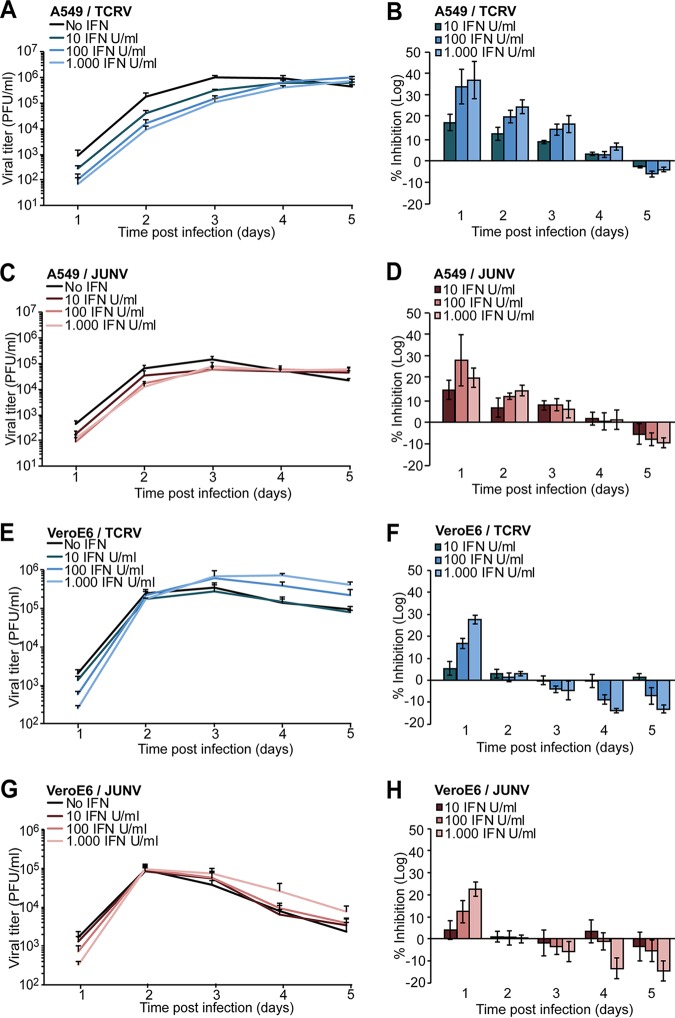
Fatal human JUNV infection is characterized by a high serum viral load and elevated IFN-I levels, suggesting limited control of the virus by IFN-I in vivo (58, 59). This was corroborated by in vitro studies that showed low sensitivity of JUNV to IFN-I treatment of human or primate cells (18). To compare the sensitivities of JUNV Candid1 and TCRV to IFN-I treatment, we exposed human A549 cells and VeroE6 cells to increasing doses of recombinant IFN-I for 20 h, followed by infection at a low MOI to allow multiple rounds of viral replication. Infectious virus production was monitored over 5 days by IFA. In A549 cells, IFN-I treatment reduced titers of JUNV Candid1 and TCRV during the first 3 days of infection in a dose-dependent manner (Fig. 3A to D). The inhibitory effects of IFN-I treatment diminished by day 4 and seemed to reverse by day 5, when we repeatedly observed slightly higher viral titers in IFN-I-treated cells (Fig. 3A to D). In VeroE6 cells, IFN-I treatment mildly reduced JUNV Candid1 titers after 1 day, with negligible effects after 2 days, which was in line with published reports (18) (Fig. 3E to H). After 3 days, JUNV Candid1 and TCRV titers from IFN-pretreated VeroE6 cells slightly surpassed viral titers from nontreated cells (Fig. 3E to H). In summary, these results indicated similar low sensitivities of TCRV and JUNV infection to IFN-I treatment.
FIG 3.
Low sensitivity of JUNV Candid1 and TCRV to IFN-I treatment. A549 (panels A to D) and VeroE6 (panels E to H) cells were preincubated for 20 h with the indicated doses of recombinant human IFN-I, followed by infection with TCRV or JUNV Candid1 at an MOI of 0.1 PFU/cell. Conditioned cell culture supernatants were collected daily over 5 days and tested for viral titer by IFA. Inhibition was calculated as 1 – [(log(mock) − log(sample))/(log(mock)], where “mock” and “sample” are viral titers of mock-infected cells and infected cells, respectively, obtained at each day postinfection. Data are means ± SD (n = 3), with one representative example of three independent experiments shown.
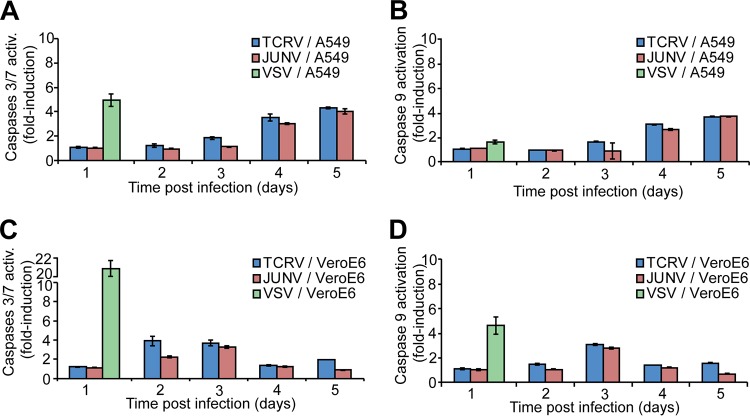
JUNV Candid1 and TCRV induce similar levels of apoptosis.
Another important mechanism of innate antiviral defense is the induction of apoptosis in response to infection (60, 61). Infection with some RNA viruses, including VSV and Sendai virus, can trigger mitochondrial apoptosis via a RIG-I/MAVS-dependent pathway involving activation of IRF3 and the proapoptotic protein Bax, resulting in autocatalytic cleavage of procaspase 9 and activation of effector caspases 3 and 7 (62, 63). Previous studies revealed induction of apoptosis by JUNV Romero and JUNV Candid1 via RIG-I in A549 and VeroE6 cells, whereas TCRV was proapoptotic in VeroE6 cells in other reports (64, 65). To directly compare the proapoptotic effects of JUNV Candid1 and TCRV, we infected VeroE6 and A549 cells with TCRV, JUNV Candid1, and VSV and monitored activation of caspase 9 and caspases 3/7 over time using a sensitive luminescence-based assay. Consistent with published reports (64, 65), infection of VeroE6 and A549 cells with JUNV Candid1, TCRV, and VSV induced activation of caspase 9 and caspases 3 and 7 (Fig. 4). At later time points, we observed for both arenaviruses stronger caspase activation in A549 cells than in the IFN-I-deficient VeroE6 cells, suggesting a possible contribution of IFN-I (Fig. 4). However, in contrast to the marked differences observed in the levels of IFN-β upregulation after TCRV and JUNV Candid1 infection (Fig. 1, Table 1), the patterns of caspase activation seemed rather similar (Fig. 4).
FIG 4.
JUNV Candid1 and TCRV induce similar levels of apoptosis. A549 (A and B) and VeroE6 cells (C and D) were infected with TCRV and JUNV Candid1 at an MOI 0.1 PFU/cell or with VSV at an MOI 1 PFU/cell. Cells were probed for activity of caspases 3/7 (A and C) and caspase 9 (B and D) over 5 days, and the results were normalized to the value for mock-infected cells. Data are means ± SD (n = 3), and one representative example of three independent experiments is shown.
Role of PKR in control of TCRV but not JUNV infection.
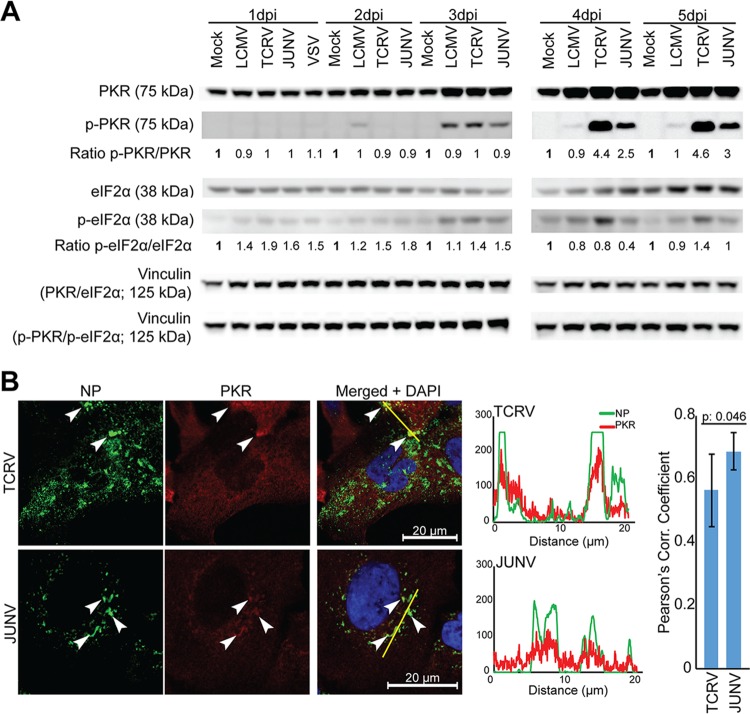
Recent studies demonstrated that JUNV and MACV activate the dsRNA-dependent PKR and that PKR colocalizes with viral replication-transcription complexes (26, 39, 40). We then investigated activation of PKR during infection with JUNV Candid1, TCRV, and LCMV. We infected monolayers of A549 cells with JUNV Candid1, TCRV, and LCMV at a low MOI. At the indicated time points, total cellular protein was extracted and subjected to Western blot analysis with specific antibodies to PKR, activated phospho-PKR, its downstream target eIF2α, and phospho-eIF2α. Vinculin served as a loading control. Infection with JUNV Candid1, TCRV, and LCMV upregulated PKR to some extent after 3 days, with stronger signals detected for the NW arenaviruses (Fig. 5A). On day 3 postinfection, JUNV Candid1, TCRV, and LCMV induced low levels of PKR phosphorylation (Fig. 5A). While phospho-PKR levels in LCMV-infected cells dropped afterwards, we consistently observed a marked increase in phospho-PKR signals at days 4 and 5 postinfection with JUNV Candid1 and TCRV (Fig. 5A). Quantification of the phospho-PKR/PKR signal ratios by densitometry consistently revealed stronger PKR phosphorylation in response to TCRV than to JUNV infection (Fig. 5A). Infection with TCRV, but not JUNV Candid1 and LCMV, resulted in detectable eIF2α phosphorylation above background on days 4 and 5 (Fig. 5A). Examination of A549 cells infected with JUNV Candid1 and TCRV by confocal microscopy revealed significant colocalization of PRK with both JUNV Candid1 and TCRV NP, indicating an association of PKR with the cytosolic replication-transcription complexes of both viruses (66) (Fig. 5B).
FIG 5.
Differential activation of PKR by JUNV Candid1 and TCRV. (A) A549 cells were infected with JUNV Candid1, TCRV, and LCMV at an MOI of 0.1 PFU/cell, and total lysates were harvested at the indicated time points (dpi, days postinfection). Total cellular proteins were probed for PKR, phospho-PKR, eIF2α, and phospho-eIF2α expression by Western blotting. Vinculin was included as a loading control in PKR/eIF2α and p-PKR/p-eIF2α membranes. The ratios of phospho-PKR/PKR (p-PKR/PKR) and phospho-eIF2α/eIF2α (p-eIF2α/eIF2α) were assessed by densitometry and normalized to the ratios of mock-infected cells at the corresponding day postinfection. One representative example out of two independent experiments is shown. (B) NP and PKR colocalization in TCRV- and JUNV Candid1-infected A549 cells. A549 cells were infected at an MOI of 1 PFU/cell, fixed, and stained 48 h postinfection with a mouse MAb to TCRV and JUNV Candid1 NP, combined with a rabbit MAb to PKR. Fluorescence intensity profiles were calculated with ImageJ/Fiji software and Pearson’s correlation coefficients with Excel software. White arrowheads, NP and PKR colocalization. Pearson’s correlation coefficient was the average result for seven analyzed cells for each virus, one of which is shown as a representative example. Bar, 20 μm.
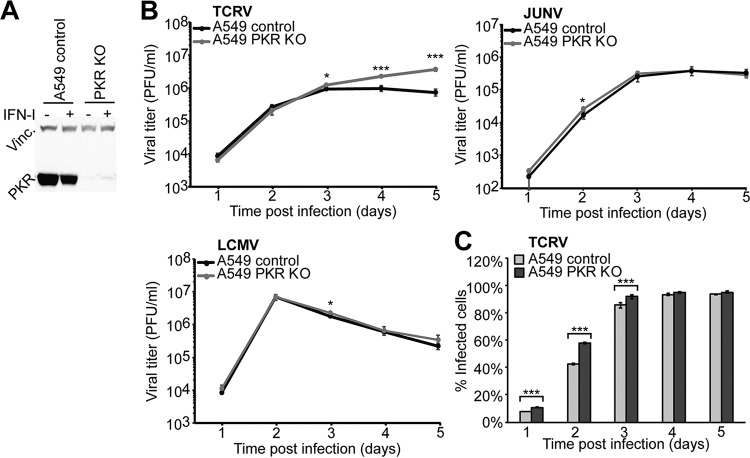
To assess a possible role of PKR in the control of TCRV and JUNV Candid1 in our system, we generated PKR-deficient A549 cells using CRISPR/Cas9 technology as described in Materials and Methods and verified deletion of PKR by Western blot analysis (Fig. 6A). Validated PKR KO and control A549 cells were infected with JUNV Candid1, TCRV, and LCMV at a low MOI, and infectious virus titers were monitored over time by IFA. Multiplication of JUNV Candid1 and LCMV were hardly affected in PKR KO cells, in line with previous studies (Fig. 6B) (39). In contrast, TCRV titers significantly increased after 4 days of infection in the absence of PKR (Fig. 6B). Finally, we monitored cell-to-cell propagation of TCRV in PKR KO and control A549 cells over time. To this end, we infected cells with TCRV (MOI, 0.1), fixed them at the time points indicated in Fig. 6 , and stained them for TCRV NP, followed by fluorescence-activated cell sorter (FACS) analysis. Consistent with the antiviral activity of PKR, we observed faster viral propagation in PKR KO A549 cells than in controls (Fig. 6C). The specific antiviral effect of PKR against TCRV, but not JUNV Candid1 or LCMV, was in line with the observed distinct activation of PKR and phosphorylation of eIF2α and suggested differential interactions of JUNV Candid1 and TCRV with PKR.
FIG 6.
PKR is involved in control of TCRV but not JUNV infection. (A) Validation of PKR deletion in A549 PKR KO cells. A549 PKR KO and control cells were incubated with or without 10,000 IFN-I IU/ml and lysed after 24 h. Lysates were probed with anti-PKR MAb and anti-vinculin MAb by Western blotting. (B) Growth kinetics of LCMV, TCRV, and JUNV Candid1 in A549 control and A549 PKR KO cells. Cells were infected with TCRV, JUNV Candid1, or LCMV at an MOI of 0.1 PFU/cell, and viral progeny production was assessed over 5 days by IFA. Data are means ± SD (n = 3), and one representative example of three independent experiments is shown. Statistical significance (t test) is indicated as follows: *, P < 0.01; **, P < 0.001; ***, P < 0.0001. (C) TCRV infection course in A549 control and A549 PKR KO cells. A549 control and A549 PKR KO cells were infected with TCRV (MOI, 0.1 PFU/cell), collected, and fixed at the indicated times postinfection, stained for TCRV NP, and analyzed by FACS. Data are means ± SD (n = 3) of the results of one representative example out of two independent experiments.
DISCUSSION
Since the 1950s, several clade B NW arenaviruses have broken the species barrier and emerged as severe human pathogens, including JUNV, MACV, GTOV, SABV, and CHAV (4). Despite striking differences in host range and virulence, genetic distances between pathogenic clade B NW arenaviruses like JUNV and MACV and some nonpathogenic clade B NW arenaviruses like TCRV are remarkably small and not well understood. Arenaviruses of clades A and C have so far not been associated with human disease. Viruses of clade D can jump between rodent species (67–72), and there is serological evidence for human infection (73), suggesting potential for spillover. Circumstantial evidence has linked the clade D White Water Arroyo virus (WWAV) to human fatal disease (74); however, a causal relationship was not established. To spill over, NW arenaviruses must undergo molecular interactions with cellular factors of the new host at different levels of their productive life cycle. These host cell factors critically contribute to the “species barrier” that emerging NW arenaviruses need to overcome.
Virus receptor use is a key determinant for zoonotic disease potential of NW arenaviruses (8–11). Structure-function analysis revealed that few point mutations suffice to convert human TfR1 into a functional receptor for a nonpathogenic NW arenavirus (8, 10, 11). A hallmark of arenaviruses is their low-fidelity mechanism of replication, resulting in high mutation rates that allow for rapid adaptation (75, 76). It is thus conceivable that currently nonpathogenic NW arenaviruses may at some point acquire the capacity to recognize human TfR1, facilitating entry into human cells. After receptor-mediated endocytosis, arenaviruses undergo low pH-dependent fusion at late endosomes (77), followed by penetration into the cytosol and initiation of viral transcription and replication. While OW arenaviruses efficiently suppress innate recognition in the cytosol by RIG-I/MAVS, NW arenaviruses seem less capable to do so (18, 27, 28). Comparative studies with recombinant proteins further revealed stronger IFN-I suppression by NP and Z of JUNV and MACV than by NP and Z of TCRV (25, 28, 37, 38), suggesting important differences between the innate responses against pathogenic and nonpathogenic NW arenaviruses, which may contribute to their distinct disease potential. To investigate this further, we undertook a systematic side-by-side comparison of the innate immune responses to JUNV Candid1 and TCRV in human cells.
Infection of human A549 cells consistently resulted in higher IFN-I levels in response to TCRV than in response to JUNV Candid1. Induction of IFN-I in response to both viruses is critically dependent on MAVS, suggesting at least overlapping pathways of innate detection. Similar intracellular levels of viral RNA observed for the two viruses made a simple correlation between IFN-I induction and viral replication level rather unlikely. Exposure of infected cells to an innate “danger signal” confirmed more efficient IFN-I suppression by JUNV than by TCRV in the infection context, consistent with previous work on recombinant NP and Z by others (28, 38). Previous studies revealed discrepancies regarding the IFN-I antagonist activity of TCRV NP, likely due to variation in amino acid residues 389 to 392 (28, 35, 78). In our study, we used TCRV strain 11573, previously used by Martinez-Sobrido et al. (28). Our data are consistent with the previously reported weaker IFN-I antagonism of NP derived from TCRV 11573 (28). Transcription profiling of 76 ISGs revealed quantitatively different, but qualitatively similar, patterns of gene regulation after challenge with TCRV and JUNV. The major ISGs induced by JUNV Candid1 and TCRV were the RIG-I, OAS, and ISG15 genes, which was in line with earlier studies with pathogenic JUNV Romero and MACV (26, 27). In summary, the data suggest recognition of JUNV and TCRV by similar innate pathways. In comparison to TCRV, JUNV seems more efficient in suppression of IFN-I induction during early infection, likely explaining the quantitative differences.
Previous studies revealed that the pathogenic JUNV Romero, JUNV Candid1, and MACV are relatively resistant to IFN-I treatment of human and primate cell lines (18, 79), which is in line with the high IFN-I levels found in severe human JUNV infection (58, 59). Here, we consistently found high levels of TCRV multiplication in the presence of a robust IFN-I response and similar low sensitivities of TCRV and JUNV to IFN-I treatment of human and primate cells. Interestingly, studies in rodent models revealed a crucial role of IFN-I in the control of JUNV, MACV, and TCRV infection in vivo and in vitro (80–84). The biological relevance of these species-specific differences is, however, currently unknown.
While the prototypic OW arenavirus LCMV induces only low levels of apoptosis in different cell lines (85), infection of A549 and VeroE6 cells with JUNV Romero and Candid1 induced apoptosis in a RIG-I-dependent, IFN-I independent manner (64). Here we observed similar levels of apoptosis induction in VeroE6 and A549 cells infected with TCRV and JUNV Candid1. Previous studies reported a role for JUNV NP as a decoy substrate for activated caspase 3 (86), providing the first evidence that arenaviruses can counteract proapoptotic signaling, an aspect that we are currently investigating for TCRV.
Recent studies revealed activation of PKR by JUNV Romero, JUNV Candid1, and MACV but not LASV (26, 39), and a recent proteomic screen revealed a specific molecular interaction between JUNV NP and PKR (39). Infection with the pathogenic JUNV Romero strain and MACV resulted in PKR-mediated phosphorylation of eIF2α that reduced early ISG translation and contributed to optimal viral multiplication (26). A similar inhibition of ISG translation by virus-induced activation of PKR was observed earlier with hepatitis C virus, which efficiently evades innate immunity and establishes long-term chronic infection in human hepatocytes (87). In contrast to pathogenic JUNV Romero, JUNV Candid1 activated PKR without inducing phosphorylation of eIF2α, and deletion of PKR had no effect on viral multiplication (39), which is consistent with our present findings. In cells infected with JUNV Candid1, phosphorylated PKR colocalized with NP and dsRNA, suggesting sequestration of the activated kinase within the viral replication-transcription complexes (39, 40). Unlike JUNV Candid1, and similar to pathogenic JUNV Romero, infection with TCRV in A549 cells activated PKR to an extent that resulted in detectable eIF2α phosphorylation. In contrast to JUNV Romero and MACV, which seem to (ab)use PKR for their own benefit, deletion of PKR enhanced production of TCRV, suggesting a role of PKR in control of the virus. Confocal microscopy revealed similar colocalizations of TCRV and JUNV NP with PKR, excluding major differences in subcellular distribution of PKR. More subtle differences, perhaps involving differential molecular interactions of JUNV NP and TCRV NP with PKR, may underlie this specific effect, a possibility we currently test.
In summary, our comparative study revealed that JUNV and TCRV induce robust IFN-I responses that differ in amplitude but result in qualitatively similar patterns of ISG expression, suggesting similar underlying mechanisms of innate recognition and signaling. Late in infection, PKR exerted some control over TCRV but not JUNV. Considering coevolution of TCRV predominantly with its reservoir host, there may be significant differences in magnitude and quality of the IFN-I response and the antiviral role of PKR between reservoir-derived and human cells. Comparative studies of TCRV infection between reservoir-derived and human cells may thus reveal differential innate responses, possibly linked to persistence in reservoirs. The current absence of serological evidence of human TCRV infection outside of laboratory accidents suggests that spillover into human populations is a rare event. This may be due to several factors, including the inability of naturally circulating TCRV variants to use human TfR1 and the antiviral action of human PKR uncovered here. However, the relative insensitivity of TCRV to IFN-I in human cells shown here suggests that once the barrier of cell entry is overcome, TCRV may be able to multiply efficiently in human cells in vivo, facilitating subsequent human adaptation.
MATERIALS AND METHODS
Antibodies and reagents.
For TCRV and JUNV detection, mouse monoclonal antibody (MAb) MA03-BE06 (88) was obtained from BEI Resources (Manassas, VA). For LCMV detection, mouse MAb 113 anti-LCMV NP was used (89). Monoclonal rabbit anti-vinculin antibody (EPR8185) was purchased from Abcam. Mouse MAb anti-MAVS was purchased from Enzo Life Sciences (Farmingdale, NY). Mouse MAb 23H12 to the M protein of VSV was kindly provided by Douglas S. Lyles (Wake Forest University School of Medicine, Winston-Salem, NC). Rabbit monoclonal antibodies against PKR (Y117) and phospho-PKR (EPR2152Y) were purchased from Abcam. Rabbit monoclonal antibodies against eIF2α (no. 9722) and phospho-eIF2α (no. 9721) were obtained from Cell Signaling Technology. Alexa Fluor 488 F(ab′)2 fragment of goat anti-mouse IgG and Alexa Fluor 594 goat anti-mouse IgG were purchased from Life Technologies (Carlsbad, CA). Polyclonal rabbit anti-mouse antibody conjugated with horseradish peroxidase (HRP) was obtained from Dako. Cell nuclei were counterstained with 4′,6′-diamidino-2-phenylindole (DAPI). Recombinant human IFN (interferon-αA/D human; catalog no. I4401) was obtained from Sigma-Aldrich.
Cells and viruses.
Human lung adenocarcinoma epithelial cells (A549), A549 MAVS KO, A549 PKR KO, human embryonic kidney cells (293T), and African green monkey kidney epithelial cells (VeroE6) were maintained in Dulbecco's modified Eagle medium containing high glucose (4.5 mg/liter) and GlutaMAX (DMEM; Gibco BRL) supplemented with 10% (vol/vol) fetal calf serum (FCS) at 37°C and 5% (vol/vol) CO2. Baby hamster kidney (BHK-21) cells were maintained in DMEM supplemented with 5% (vol/vol) FCS and nonessential amino acids (Gibco BRL) at 37°C and 5% (vol/vol) CO2. All cells were regularly tested for mycoplasma contamination with a MycoAlert mycoplasma detection kit (Lonza). To infect cells, we used Tacaribe virus (TCRV) strain 11573, Junin virus (JUNV) strain Candid1, lymphocytic choriomeningitis virus (LCMV) ARM53b variant clone 13, and vesicular stomatitis virus (VSV) serotype Indiana. TCRV and JUNV were plaque purified as described below. For the use of VSV at the Institute of Microbiology of Lausanne University Hospital, we obtained permission (no. A141238) from the Office Fédéral de la Securité Alimentaire et des Affaires Veterinaries (OSAV).
Infections, plaque purification, and viral stock production.
For viral infections, cells were seeded before infection to reach >80% confluence at infection. Viruses were then diluted in complete DMEM (500 μl for 6-well plate format; 150 μl for 24-well plate format) to obtain the desired MOI and applied to cells for 90 min at 37°C (adsorption). After adsorption, the inoculum was removed and replaced by fresh complete DMEM. For plaque purification, TCRV and JUNV were used to infect VeroE6 monolayers as described above. After adsorption, semisolid medium (colorless DMEM supplemented with 0.5% agar and 2% FCS) was gently added to each well. After 7 days, plaques were visualized under a microscope and 10 μl of medium was taken, diluted in 150 μl of complete DMEM, and used as the inoculum in fresh VeroE6 cells (24-well format) for viral propagation. For viral stock production, three passages in VeroE6 cells were performed at an MOI of 0.01 PFU/cell in increasing format flasks, with collection of supernatants after 4 days postinfection. Final amplification was performed in BHK-21 cells (HYPERflask format; Corning). Supernatants were cleared by centrifugation at 1,500 rpm for 5 min at 4°C. Cleared samples were mixed 1:1 with a sterile solution of 140 g/liter polyethylene 8000 (PEG8000) in phosphate-buffered saline (PBS) and incubated overnight under shaking. Samples were centrifuged at 8,000 × g for 1 h at 4°C. Supernatants were discarded, and pellets were resuspended in complete DMEM. PEG-precipitated samples were then layered on top of a cushion of 30% (wt/vol) sucrose in PBS and centrifuged at 20,000 rpm for 2 h at 4°C in an Optima XPN-80 ultracentrifuge (Beckman Coulter) with a SW-55 Ti rotor. After ultracentrifugation, pellets were resuspended in complete DMEM and stored at −80°C.
Fluorescence-activated cell sorting (FACS).
Cells were detached, resuspended, and washed once with complete DMEM, washed once with PBS, and fixed with 2% (wt/vol) formaldehyde in PBS for 30 min at room temperature. After washing with PBS, cells were permeabilized for 30 min at room temperature with PBS supplemented with 0.1% (wt/vol) saponin and 1% (vol/vol) FCS in PBS. Primary and secondary antibodies were diluted in permeabilization solution and incubated for 1 h and 45 min, respectively, at room temperature. Cells were finally washed three times with PBS and analyzed with a FACSCalibur flow cytometer (Becton, Dickinson, San Jose, CA).
Immunofocus assay (IFA).
VeroE6 cells were seeded at 1.5 × 104 cells/well in 96-well plates and infected on the following day with 10-fold serial dilutions of the examined samples. After adsorption, inocula were removed and 100 μl of fresh medium was added to each well. At 16 to 20 h after infection, cells were washed once with PBS and fixed with 2% (wt/vol) formaldehyde in PBS for 30 min at room temperature. Cells were washed with PBS and permeabilized for 30 min at room temperature with 0.1% (wt/vol) saponin and 1% (vol/vol) FCS in PBS. Primary and secondary antibodies were diluted with permeabilization solution and incubated for 1 h and 45 min, respectively, at room temperature. Cells were washed three times with PBS. Positive infectious foci were scored using an EVOS FLoid cell imaging station with a 20× Plan fluorite lens (Thermo Fisher).
IFN-I bioassay.
To detect the biological activity of IFN-I produced by A549 cells infected with TCRV or JUNV Candid1, we performed an IFN-I bioassay as described previously (28). Briefly, we treated VeroE6 cells with tissue culture supernatants derived from TCRV- or JUNV Candid1-infected A549 cells that had been previously subjected to UV treatment to inactivate infectious viral particles. UV treatment was performed at 4°C, in a rocking station at 10 cm from the UV irradiation source. After 16 h of incubation, we examined the susceptibility of these cells to infection by VSV, which is known to be highly susceptible to IFN-I (90), using IFA against the VSV M protein, as reported previously (85).
RNA isolation, RT-qPCR, and transcriptome profiler.
Total RNA was isolated with a NucleoSpin RNA kit (Macherey-Nagel) and eluted in 60 μl of water, in accordance with the manufacturer’s instructions, or with an RNeasy mini kit (Qiagen) performed by the EZ1 Advanced XL robot at the service of Molecular Diagnostics of the Institute of Microbiology, University of Lausanne, and eluted in 50 μl of water. Total RNA was quantified with NanoDrop 1000 or Qubit 4.0 (Thermo Fisher), by following the manufacturer’s instructions. Five hundred nanograms of total RNA was used for reverse transcription reactions using a high-capacity cDNA reverse transcription kit from Applied Biosystems (Foster City, CA), in accordance with the manufacturer’s protocol. TaqMan probes specific for human IFN-β (Hs01077958_s1/FAM) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH; Hs99999905_m1/VIC) were purchased from Applied Biosystems. TaqMan probes targeting TCRV NP (For, 5′-TGCACAGTGAGGTTCATCAG-3′; Rev, 5′-GAGCACCAAGTGTGGGATAA-3′; and probe, 5′-6FAM-TATGTTGTTTGCAGAGGTGGCCGABHQ1-3′) and JUNV NP (For, 5′-CATGGAGGTCAAACAACTTCCT-3′; Rev, 5′-GCCTCCAGACATGGTTGTGA-3′; and probe, 5′-6FAM-ATGTCATCGGATCCTTMGBNFQ-3′) were obtained from Thermo Fisher. Quantitative PCR (qPCR) was performed using a StepOne qPCR system (Applied Biosystems), and gene expression levels relative to GAPDH were determined according to the ΔΔCT (where CT is threshold cycle) method (91). For transcriptome profiling, human antiviral response RT2 Profiler PCR array kits (PAHS-122ZG; Qiagen) were used. A total of 3.5 μg (quantified by Qubit 4.0) of total cellular RNA was used for the reverse transcription reaction (RT2 SYBR green qPCR mastermix; Qiagen). RT2 Profiler PCR array 384-well plates were set up by a PIRO personal pipetting robot. All RNA samples were tested for quality in a fragment analyzer (Agilent), selecting only those with an RNA quality score (RIN) of 10. All tested samples were negative for genomic DNA contamination. Samples with aberrant amplification curves or shifted or multiple melting peaks were discarded from the analysis. qPCR was done in a LightCycler 480 instrument II in accordance with the manufacturer’s instructions.
Detection of caspase activity.
To monitor the activation of caspases 9 and 3/7, we used the Caspase-Glo 9 and 3/7 assay kits from Promega (Madison, WI). Briefly, 2 × 104 A549 cells or 1.5 × 104 VeroE6 cells per well were seeded in 96-well white clear-bottom plates on the day prior to the experiment. Cells were then infected with viruses and processed according to the manufacturer’s protocol, and caspase activity was measured by luminescence detection in a TriStar LB 941 multimode microplate reader from Berthold Technologies (Bad Wildbad, Germany).
Transfections and plasmids.
Transfections in 293T cells were performed with a jetPRIME transfection kit, using 0.55 μg of total DNA per well (24-well plate format). Transfection efficiency control was performed in parallel with the pcDNA3.1 plasmid encoding enhanced green fluorescent protein (EGFP). pIFN-LUC plasmid was kindly provided by Verónica Martin and encodes firefly luciferase under the control of type I interferon promoter (ISRE).
Luciferase and CellTiter-Glo assays.
For luminescence assays, cells were washed once with PBS and assayed for luciferase activity using the Luciferase assay system (Promega) or Dual-Luciferase reporter assay system (Promega). Cell viability was used to normalize luciferase-based assays. Cell monolayers were washed once, and PBS was added to the cells. Cells were then processed for the CellTiter-Glo assay (Promega) as indicated by the manufacturer and read in a TriStar LB 941 multimode microplate reader from Berthold Technologies (Bad Wildbad, Germany).
Generation of CRISPR/Cas9 knockout cell lines.
A549 MAVS KO and PKR KO cells were generated by using the CRISPR/Cas9 technology. Briefly, single guide RNA (sgRNA) sequences targeting the exon regions of PKR (5′-CAGGACCTCCACATGATAGG-3′) and MAVS (5′-CCGACCGGAAGTTCCAGGAG-3′) were selected using CHOPCHOP (1), a web tool for genome editing. A nontargeting sgRNA (5′-CTAAGGTTAAGTCGCCCTCG-3′) was used as a control. Annealed oligonucleotides were cloned into pLenti CRISPR v2 ccdB (2, 3) via a single-step Golden Gate cloning approach. In order to transduce A549 cells with the respective gRNA sequence, lentiviral VSV-G pseudotyped particles were produced in the cells. Therefore, 293T cells were transfected with plasmid DNA encoding the human immune deficiency virus (HIV) Gag and Pol proteins, VSV-G protein, and the respective pLenti CRISPR construct. Lentiviruses were harvested 48 h posttransfection, and A549 cells were transduced with the lentiviruses for 8 h. At 48 h postransduction, cells were subjected to puromycin selection at a concentration of 2 μg/ml. The batch knockout cell population was further diluted by limiting dilutions and seeded at ∼0.5 to 2 cells/well in 96-well plates, and single-cell clones were expanded for 3 weeks. Clones were characterized by immunoblotting.
Immunoblotting.
For immunoblotting, proteins were separated by SDS-PAGE and transferred to nitrocellulose. After blocking in 5% (wt/vol) skim milk in PBS, membranes were incubated with 1 to 10 μg/ml primary antibody in 5% (wt/vol) skim milk–PBS overnight at 4°C. After several washes in PBS containing 0.1% (wt/vol) Tween 20 (PBST), secondary antibodies coupled to HRP were applied at 1:3,000 in PBST for 1 h at room temperature. Membranes were developed by enhanced chemiluminescence (ECL) using a LiteABlot kit (EuroClone). Signals were acquired by a ImageQuant LAS 4000 Mini (GE Healthcare Lifesciences). Quantification of Western blot results was performed with ImageJ software.
Confocal microscopy.
To visualize viral NP (from TCRV, JUNV, and LCMV) in double staining with PKR, A549 cells were seeded on 12-mm coverslips and infected with viruses as indicated in the figures. Specimens were fixed with 2% (wt/vol) formaldehyde in PBS for 30 min at room temperature and washed with PBS. Cells were then permeabilized for 30 min at room temperature with PBS supplemented with 0.1% (wt/vol) saponin and 1% (vol/vol) FCS. Primary and secondary antibodies were diluted in permeabilization solution and incubated overnight at 4°C and for 1 h at room temperature, respectively. Cell nuclei were counterstained with 4′,6′-diamidino-2-phenylindole (DAPI). Specimens were finally mounted using Mowiol (Sigma-Aldrich, St. Louis, MO). Image acquisition was performed with a Zeiss LSM780 Quasar confocal microscope. Multiplier gain for each channel was adjusted to minimize background noise and saturated pixels. To quantify colocalization, cells were examined by measuring fluorescence intensity (ImageJ/Fiji software) along a line >12 μm long. Pearson’s correlation coefficients were calculated from fluorescence intensity values of PKR or phospho-PKR and viral NP obtained from all examined cells.
ACKNOWLEDGMENTS
We thank Juan Carlos de la Torre (Scripps Research Institute, La Jolla, CA, USA) for the TCRV isolate, Michael Buchmeier (Department of Molecular Biology and Biochemistry, University of California, Irvine, CA, USA) for the Candid1 vaccine strain, Marc Schmidt-Supprian (Department of Hematology and Medical Oncology, Munich, Germany) and Klaus Heger (Department of Physiological Chemistry, Genentech, South San Francisco, CA, USA) for the pLenti CRISPR v2 ccdB plasmid, and Veronica Martin for the pIFN-LUC plasmid (Department of Molecular and Cell Biology, Centro Nacional de Biotecnología, Madrid, Spain). The following reagent was obtained through BEI Resources, NIAID, NIH: monoclonal anti-Junin virus, clone MA03-BE06, NR-41860.
This research was supported by Swiss National Science Foundation grants SINERGIA Nr. CRSII3_160780/1 and 310030_170108 to S.K. and funds to S.K. from the University of Lausanne. This work was further supported by grants from the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation)—Projektnummer 158989968 (SFB900, project C7; http://www.sfb900.de), the Knut and Alice Wallenberg Foundation, and the German Liver Foundation (S163/10135/2017) to G.G. This work was also supported by a fellowship from the Hanover Biomedical Research School and the Centre for Infection Biology (ZIB; https://www.mh-hannover.de/zib_zib.html) to R.M.
REFERENCES
- 1.Buchmeier MJ, de la Torre JC, Peters CJ. 2007. Arenaviridae: the viruses and their replication, p 1791–1828. In Knipe DL, Howley PM (ed), Fields virology, 4th ed Lippincott-Raven, Philadelphia, PA. [Google Scholar]
- 2.Geisbert TW, Jahrling PB. 2004. Exotic emerging viral diseases: progress and challenges. Nat Med 10:S110–S1121. doi: 10.1038/nm1142. [DOI] [PubMed] [Google Scholar]
- 3.Radoshitzky SR, Bao Y, Buchmeier MJ, Charrel RN, Clawson AN, Clegg CS, DeRisi JL, Emonet S, Gonzalez JP, Kuhn JH, Lukashevich IS, Peters CJ, Romanowski V, Salvato MS, Stenglein MD, de la Torre JC. 2015. Past, present, and future of arenavirus taxonomy. Arch Virol 160:1851–1874. doi: 10.1007/s00705-015-2418-y. [DOI] [PubMed] [Google Scholar]
- 4.Sarute N, Ross SR. 2017. New World arenavirus biology. Annu Rev Virol 4:141–158. doi: 10.1146/annurev-virology-101416-042001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de la Torre JC. 2009. Molecular and cell biology of the prototypic arenavirus LCMV: implications for understanding and combating hemorrhagic fever arenaviruses. Ann N Y Acad Sci 1171(Suppl 1):E57–E64. doi: 10.1111/j.1749-6632.2009.05048.x. [DOI] [PubMed] [Google Scholar]
- 6.Peters CJ. 2002. Human infection with arenaviruses in the Americas. Curr Top Microbiol Immunol 262:65–74. [DOI] [PubMed] [Google Scholar]
- 7.Borio L, Inglesby T, Peters CJ, Schmaljohn AL, Hughes JM, Jahrling PB, Ksiazek T, Johnson KM, Meyerhoff A, O'Toole T, Ascher MS, Bartlett J, Breman JG, Eitzen EM Jr, Hamburg M, Hauer J, Henderson DA, Johnson RT, Kwik G, Layton M, Lillibridge S, Nabel GJ, Osterholm MT, Perl TM, Russell P, Tonat K. 2002. Hemorrhagic fever viruses as biological weapons: medical and public health management. JAMA 287:2391–2405. doi: 10.1001/jama.287.18.2391. [DOI] [PubMed] [Google Scholar]
- 8.Abraham J, Kwong JA, Albarino CG, Lu JG, Radoshitzky SR, Salazar-Bravo J, Farzan M, Spiropoulou CF, Choe H. 2009. Host-species transferrin receptor 1 orthologs are cellular receptors for nonpathogenic new world clade B arenaviruses. PLoS Pathog 5:e1000358. doi: 10.1371/journal.ppat.1000358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Radoshitzky SR, Abraham J, Spiropoulou CF, Kuhn JH, Nguyen D, Li W, Nagel J, Schmidt PJ, Nunberg JH, Andrews NC, Farzan M, Choe H. 2007. Transferrin receptor 1 is a cellular receptor for New World haemorrhagic fever arenaviruses. Nature 446:92–96. doi: 10.1038/nature05539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Radoshitzky SR, Kuhn JH, Spiropoulou CF, Albarino CG, Nguyen DP, Salazar-Bravo J, Dorfman T, Lee AS, Wang E, Ross SR, Choe H, Farzan M. 2008. Receptor determinants of zoonotic transmission of New World hemorrhagic fever arenaviruses. Proc Natl Acad Sci U S A 105:2664–2669. doi: 10.1073/pnas.0709254105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zong M, Fofana I, Choe H. 2014. Human and host species transferrin receptor 1 use by North American arenaviruses. J Virol 88:9418–9428. doi: 10.1128/JVI.01112-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jemielity S, Wang JJ, Chan YK, Ahmed AA, Li W, Monahan S, Bu X, Farzan M, Freeman GJ, Umetsu DT, Dekruyff RH, Choe H. 2013. TIM-family proteins promote infection of multiple enveloped viruses through virion-associated phosphatidylserine. PLoS Pathog 9:e1003232. doi: 10.1371/journal.ppat.1003232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Amara A, Mercer J. 2015. Viral apoptotic mimicry. Nat Rev Microbiol 13:461–469. doi: 10.1038/nrmicro3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Borrow P, Martinez-Sobrido L, de la Torre JC. 2010. Inhibition of the type I interferon antiviral response during arenavirus infection. Viruses 2:2443–2480. doi: 10.3390/v2112443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koma T, Huang C, Kolokoltsova OA, Brasier AR, Paessler S. 2013. Innate immune response to arenaviral infection: a focus on the highly pathogenic New World hemorrhagic arenaviruses. J Mol Biol 425:4893–4903. doi: 10.1016/j.jmb.2013.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meyer B, Ly H. 2016. Inhibition of innate immune responses is key to pathogenesis by arenaviruses. J Virol 90:3810–3818. doi: 10.1128/JVI.03049-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shao J, Liang Y, Ly H. 2015. Human hemorrhagic fever causing arenaviruses: molecular mechanisms contributing to virus virulence and disease pathogenesis. Pathogens 4:283–306. doi: 10.3390/pathogens4020283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang C, Kolokoltsova OA, Yun NE, Seregin AV, Poussard AL, Walker AG, Brasier AR, Zhao Y, Tian B, de la Torre JC, Paessler S. 2012. Junin virus infection activates the type I interferon pathway in a RIG-I-dependent manner. PLoS Negl Trop Dis 6:e1659. doi: 10.1371/journal.pntd.0001659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kato H, Takahasi K, Fujita T. 2011. RIG-I-like receptors: cytoplasmic sensors for non-self RNA. Immunol Rev 243:91–98. doi: 10.1111/j.1600-065X.2011.01052.x. [DOI] [PubMed] [Google Scholar]
- 20.Yoneyama M, Onomoto K, Jogi M, Akaboshi T, Fujita T. 2015. Viral RNA detection by RIG-I-like receptors. Curr Opin Immunol 32:48–53. doi: 10.1016/j.coi.2014.12.012. [DOI] [PubMed] [Google Scholar]
- 21.Vazquez C, Horner SM. 2015. MAVS coordination of antiviral innate immunity. J Virol 89:6974–6977. doi: 10.1128/JVI.01918-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kell AM, Gale M Jr. 2015. RIG-I in RNA virus recognition. Virology 479-480:110–121. doi: 10.1016/j.virol.2015.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chan YK, Gack MU. 2015. RIG-I-like receptor regulation in virus infection and immunity. Curr Opin Virol 12:7–14. doi: 10.1016/j.coviro.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martínez-Sobrido L, Zúñiga EI, Rosario D, García-Sastre A, de la Torre JC. 2006. Inhibition of the type I interferon response by the nucleoprotein of the prototypic arenavirus lymphocytic choriomeningitis virus. J Virol 80:9192–9199. doi: 10.1128/JVI.00555-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rodrigo WWSI, Ortiz-Riaño E, Pythoud C, Kunz S, de la Torre JC, Martínez-Sobrido L. 2012. Arenavirus nucleoproteins prevent activation of nuclear factor kappa B. J Virol 86:8185–8197. doi: 10.1128/JVI.07240-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang C, Kolokoltsova OA, Mateer EJ, Koma T, Paessler S. 2017. Highly pathogenic New World arenavirus infection activates the pattern recognition receptor protein kinase R without attenuating virus replication in human cells. J Virol 91:e01090-17. doi: 10.1128/JVI.01090-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang C, Kolokoltsova OA, Yun NE, Seregin AV, Ronca S, Koma T, Paessler S. 2015. Highly pathogenic New World and Old World human arenaviruses induce distinct interferon responses in human cells. J Virol 89:7079–7088. doi: 10.1128/JVI.00526-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martínez-Sobrido L, Giannakas P, Cubitt B, García-Sastre A, de la Torre JC. 2007. Differential inhibition of type I interferon induction by arenavirus nucleoproteins. J Virol 81:12696–12703. doi: 10.1128/JVI.00882-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cuevas CD, Lavanya M, Wang E, Ross SR. 2011. Junin virus infects mouse cells and induces innate immune responses. J Virol 85:11058–11068. doi: 10.1128/JVI.05304-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hastie KM, Bale S, Kimberlin CR, Saphire EO. 2012. Hiding the evidence: two strategies for innate immune evasion by hemorrhagic fever viruses. Curr Opin Virol 2:151–156. doi: 10.1016/j.coviro.2012.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martínez-Sobrido L, Emonet S, Giannakas P, Cubitt B, García-Sastre A, de la Torre JC. 2009. Identification of amino acid residues critical for the anti-interferon activity of the nucleoprotein of the prototypic arenavirus lymphocytic choriomeningitis virus. J Virol 83:11330–11340. doi: 10.1128/JVI.00763-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qi X, Lan S, Wang W, Schelde LM, Dong H, Wallat GD, Ly H, Liang Y, Dong C. 2010. Cap binding and immune evasion revealed by Lassa nucleoprotein structure. Nature 468:779–783. doi: 10.1038/nature09605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hastie KM, Kimberlin CR, Zandonatti MA, MacRae IJ, Saphire EO. 2011. Structure of the Lassa virus nucleoprotein reveals a dsRNA-specific 3' to 5' exonuclease activity essential for immune suppression. Proc Natl Acad Sci U S A 108:2396–2401. doi: 10.1073/pnas.1016404108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y, Li L, Liu X, Dong S, Wang W, Huo T, Guo Y, Rao Z, Yang C. 2013. Crystal structure of Junin virus nucleoprotein. J Gen Virol 94:2175–2183. doi: 10.1099/vir.0.055053-0. [DOI] [PubMed] [Google Scholar]
- 35.Jiang X, Huang Q, Wang W, Dong H, Ly H, Liang Y, Dong C. 2013. Structures of arenaviral nucleoproteins with triphosphate dsRNA reveal a unique mechanism of immune suppression. J Biol Chem 288:16949–16959. doi: 10.1074/jbc.M112.420521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang Q, Shao J, Lan S, Zhou Y, Xing J, Dong C, Liang Y, Ly H. 2015. In vitro and in vivo characterizations of pichinde viral nucleoprotein exoribonuclease functions. J Virol 89:6595–6607. doi: 10.1128/JVI.00009-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fan L, Briese T, Lipkin WI. 2010. Z proteins of New World arenaviruses bind RIG-I and interfere with type I interferon induction. J Virol 84:1785–1791. doi: 10.1128/JVI.01362-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xing J, Ly H, Liang Y. 2015. The Z proteins of pathogenic but not nonpathogenic arenaviruses inhibit the RIG-I-like receptor (RLR)-dependent interferon production. J Virol 89:2944–2955. doi: 10.1128/JVI.03349-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.King BR, Hershkowitz D, Eisenhauer PL, Weir ME, Ziegler CM, Russo J, Bruce EA, Ballif BA, Botten J. 2017. A map of the arenavirus nucleoprotein-host protein interactome reveals that Junin virus selectively impairs the antiviral activity of double-stranded RNA-activated protein kinase (PKR). J Virol 91:e00763-17. doi: 10.1128/JVI.00763-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mateer EJ, Paessler S, Huang C. 2018. Visualization of double-stranded RNA colocalizing with pattern recognition receptors in arenavirus infected cells. Front Cell Infect Microbiol 8:251. doi: 10.3389/fcimb.2018.00251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Enria DA, Barrera Oro JG. 2002. Junin virus vaccines. Curr Top Microbiol Immunol 263:239–261. [DOI] [PubMed] [Google Scholar]
- 42.Albarino CG, Bird BH, Chakrabarti AK, Dodd KA, Flint M, Bergeron E, White DM, Nichol ST. 2011. The major determinant of attenuation in mice of the Candid1 vaccine for Argentine hemorrhagic fever is located in the G2 glycoprotein transmembrane domain. J Virol 85:10404–10408. doi: 10.1128/JVI.00856-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Seregin AV, Yun NE, Miller M, Aronson J, Smith JK, Walker AG, Smith JN, Huang C, Manning JT, de la Torre JC, Paessler S. 2015. The glycoprotein precursor gene of Junin virus determines the virulence of the Romero strain and the attenuation of the Candid #1 strain in a representative animal model of Argentine hemorrhagic fever. J Virol 89:5949–5956. doi: 10.1128/JVI.00104-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goni SE, Iserte JA, Stephan BI, Borio CS, Ghiringhelli PD, Lozano ME. 2010. Molecular analysis of the virulence attenuation process in Junin virus vaccine genealogy. Virus Genes 40:320–328. doi: 10.1007/s11262-010-0450-2. [DOI] [PubMed] [Google Scholar]
- 45.Pythoud C, Rodrigo WW, Pasqual G, Rothenberger S, Martinez-Sobrido L, de la Torre JC, Kunz S. 2012. Arenavirus nucleoprotein targets interferon regulatory factor-activating kinase IKKε. J Virol 86:7728–7738. doi: 10.1128/JVI.00187-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meertens L, Carnec X, Lecoin MP, Ramdasi R, Guivel-Benhassine F, Lew E, Lemke G, Schwartz O, Amara A. 2012. The TIM and TAM families of phosphatidylserine receptors mediate dengue virus entry. Cell Host Microbe 12:544–557. doi: 10.1016/j.chom.2012.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Herrador A, Fedeli C, Radulovic E, Campbell KP, Moreno H, Gerold G, Kunz S. 2019. Dynamic dystroglycan complexes mediate cell entry of Lassa virus. mBio 10:e02869-18. doi: 10.1128/mBio.02869-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Samuel CE. 2001. Antiviral actions of interferons. Clin Microbiol Rev 14:778–809. doi: 10.1128/CMR.14.4.778-809.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou S, Cerny AM, Zacharia A, Fitzgerald KA, Kurt-Jones EA, Finberg RW. 2010. Induction and inhibition of the type I IFN responses by distinct components of lymphocytic choriomeningitis virus (LCMV). J Virol 84:9452–9462. doi: 10.1128/JVI.00155-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager R, Tschopp J. 2005. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- 51.Chiu YH, Macmillan JB, Chen ZJ. 2009. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell 138:576–591. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen Q, Sun L, Chen ZJ. 2016. Regulation and function of the cGAS-STING pathway of cytosolic DNA sensing. Nat Immunol 17:1142–1149. doi: 10.1038/ni.3558. [DOI] [PubMed] [Google Scholar]
- 53.Zevini A, Olagnier D, Hiscott J. 2017. Crosstalk between cytoplasmic RIG-I and STING sensing pathways. Trends Immunol 38:194–205. doi: 10.1016/j.it.2016.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yoneyama M, Kikuchi M, Matsumoto K, Imaizumi T, Miyagishi M, Taira K, Foy E, Loo YM, Gale M Jr, Akira S, Yonehara S, Kato A, Fujita T. 2005. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J Immunol 175:2851–2858. doi: 10.4049/jimmunol.175.5.2851. [DOI] [PubMed] [Google Scholar]
- 55.Satoh T, Kato H, Kumagai Y, Yoneyama M, Sato S, Matsushita K, Tsujimura T, Fujita T, Akira S, Takeuchi O. 2010. LGP2 is a positive regulator of RIG-I- and MDA5-mediated antiviral responses. Proc Natl Acad Sci U S A 107:1512–1517. doi: 10.1073/pnas.0912986107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Venkataraman T, Valdes M, Elsby R, Kakuta S, Caceres G, Saijo S, Iwakura Y, Barber GN. 2007. Loss of DExD/H box RNA helicase LGP2 manifests disparate antiviral responses. J Immunol 178:6444–6455. doi: 10.4049/jimmunol.178.10.6444. [DOI] [PubMed] [Google Scholar]
- 57.Suthar MS, Ramos HJ, Brassil MM, Netland J, Chappell CP, Blahnik G, McMillan A, Diamond MS, Clark EA, Bevan MJ, Gale M Jr.. 2012. The RIG-I-like receptor LGP2 controls CD8(+) T cell survival and fitness. Immunity 37:235–248. doi: 10.1016/j.immuni.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Levis SC, Saavedra MC, Ceccoli C, Falcoff E, Feuillade MR, Enria DA, Maiztegui JI, Falcoff R. 1984. Endogenous interferon in Argentine hemorrhagic fever. J Infect Dis 149:428–433. doi: 10.1093/infdis/149.3.428. [DOI] [PubMed] [Google Scholar]
- 59.Levis SC, Saavedra MC, Ceccoli C, Feuillade MR, Enria DA, Maiztegui JI, Falcoff R. 1985. Correlation between endogenous interferon and the clinical evolution of patients with Argentine hemorrhagic fever. J Interferon Res 5:383–389. doi: 10.1089/jir.1985.5.383. [DOI] [PubMed] [Google Scholar]
- 60.Barber GN. 2001. Host defense, viruses and apoptosis. Cell Death Differ 8:113–126. doi: 10.1038/sj.cdd.4400823. [DOI] [PubMed] [Google Scholar]
- 61.Boatright KM, Salvesen GS. 2003. Mechanisms of caspase activation. Curr Opin Cell Biol 15:725–731. doi: 10.1016/j.ceb.2003.10.009. [DOI] [PubMed] [Google Scholar]
- 62.Chattopadhyay S, Marques JT, Yamashita M, Peters KL, Smith K, Desai A, Williams BR, Sen GC. 2010. Viral apoptosis is induced by IRF-3-mediated activation of Bax. EMBO J 29:1762–1773. doi: 10.1038/emboj.2010.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chattopadhyay S, Yamashita M, Zhang Y, Sen GC. 2011. The IRF-3/Bax-mediated apoptotic pathway, activated by viral cytoplasmic RNA and DNA, inhibits virus replication. J Virol 85:3708–3716. doi: 10.1128/JVI.02133-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kolokoltsova OA, Grant AM, Huang C, Smith JK, Poussard AL, Tian B, Brasier AR, Peters CJ, Tseng CT, de la Torre JC, Paessler S. 2014. RIG-I enhanced interferon independent apoptosis upon Junin virus infection. PLoS One 9:e99610. doi: 10.1371/journal.pone.0099610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wolff S, Groseth A, Meyer B, Jackson D, Strecker T, Kaufmann A, Becker S. 2016. The New World arenavirus Tacaribe virus induces caspase-dependent apoptosis in infected cells. J Gen Virol 97:855–866. doi: 10.1099/jgv.0.000403. [DOI] [PubMed] [Google Scholar]
- 66.Baird NL, York J, Nunberg JH. 2012. Arenavirus infection induces discrete cytosolic structures for RNA replication. J Virol 86:11301–11310. doi: 10.1128/JVI.01635-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fulhorst CF, Charrel RN, Weaver SC, Ksiazek TG, Bradley RD, Milazzo ML, Tesh RB, Bowen MD. 2001. Geographic distribution and genetic diversity of Whitewater Arroyo virus in the southwestern United States. Emerg Infect Dis 7:403–407. doi: 10.3201/eid0703.010306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cajimat MN, Milazzo ML, Hess BD, Rood MP, Fulhorst CF. 2007. Principal host relationships and evolutionary history of the North American arenaviruses. Virology 367:235–243. doi: 10.1016/j.virol.2007.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Milazzo ML, Cajimat MN, Haynie ML, Abbott KD, Bradley RD, Fulhorst CF. 2008. Diversity among Tacaribe serocomplex viruses (family Arenaviridae) naturally associated with the white-throated woodrat (Neotoma albigula) in the southwestern United States. Vector Borne Zoonotic Dis 8:523–540. doi: 10.1089/vbz.2007.0239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cajimat MN, Milazzo ML, Haynie ML, Hanson JD, Bradley RD, Fulhorst CF. 2011. Diversity and phylogenetic relationships among the North American Tacaribe serocomplex viruses (family Arenaviridae). Virology 421:87–95. doi: 10.1016/j.virol.2011.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cajimat MN, Milazzo ML, Mauldin MR, Bradley RD, Fulhorst CF. 2013. Diversity among Tacaribe serocomplex viruses (family Arenaviridae) associated with the southern plains woodrat (Neotoma micropus). Virus Res 178:486–494. doi: 10.1016/j.virusres.2013.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Milazzo ML, Cajimat MN, Mauldin MR, Bennett SG, Hess BD, Rood MP, Conlan CA, Nguyen K, Wekesa JW, Ramos RD, Bradley RD, Fulhorst CF. 2015. Epizootiology of Tacaribe serocomplex viruses (Arenaviridae) associated with neotomine rodents (Cricetidae, Neotominae) in southern California. Vector Borne Zoonotic Dis 15:156–166. doi: 10.1089/vbz.2014.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Milazzo ML, Campbell GL, Fulhorst CF. 2011. Novel arenavirus infection in humans, United States. Emerg Infect Dis 17:1417–1420. doi: 10.3201/eid1708.110285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Centers for Disease Control and Prevention. 2000. Fatal illnesses associated with a new world arenavirus–California, 1999-2000. MMWR Morb Mortal Wkly Rep 49:709–711. [PubMed] [Google Scholar]
- 75.Sevilla N, de la Torre JC. 2006. Arenavirus diversity and evolution: quasispecies in vivo. Curr Top Microbiol Immunol 299:315–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Emonet SF, de la Torre JC, Domingo E, Sevilla N. 2009. Arenavirus genetic diversity and its biological implications. Infect Genet Evol 9:417–429. doi: 10.1016/j.meegid.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fedeli C, Moreno H, Kunz S. 2018. Novel insights into cell entry of emerging human pathogenic arenaviruses. J Mol Biol 430:1839–1852. doi: 10.1016/j.jmb.2018.04.026. [DOI] [PubMed] [Google Scholar]
- 78.Harmon B, Kozina C, Maar D, Carpenter TS, Branda CS, Negrete OA, Carson BD. 2013. Identification of critical amino acids within the nucleoprotein of Tacaribe virus important for anti-interferon activity. J Biol Chem 288:8702–8711. doi: 10.1074/jbc.M112.444760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Canonico PG, Kende M, Luscri BJ, Huggins JW. 1984. In-vivo activity of antivirals against exotic RNA viral infections. J Antimicrob Chemother 14(Suppl A):27–41. doi: 10.1093/jac/14.suppl_a.27. [DOI] [PubMed] [Google Scholar]
- 80.Kolokoltsova OA, Yun NE, Poussard AL, Smith JK, Smith JN, Salazar M, Walker A, Tseng CT, Aronson JF, Paessler S. 2010. Mice lacking alpha/beta and gamma interferon receptors are susceptible to Junin virus infection. J Virol 84:13063–13067. doi: 10.1128/JVI.01389-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Huang C, Walker AG, Grant AM, Kolokoltsova OA, Yun NE, Seregin AV, Paessler S. 2014. Potent inhibition of Junin virus infection by interferon in murine cells. PLoS Negl Trop Dis 8:e2933. doi: 10.1371/journal.pntd.0002933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Patterson M, Seregin A, Huang C, Kolokoltsova O, Smith J, Miller M, Smith J, Yun N, Poussard A, Grant A, Tigabu B, Walker A, Paessler S. 2014. Rescue of a recombinant Machupo virus from cloned cDNAs and in vivo characterization in interferon (alphabeta/gamma) receptor double knockout mice. J Virol 88:1914–1923. doi: 10.1128/JVI.02925-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pedras-Vasconcelos JA, Goucher D, Puig M, Tonelli LH, Wang V, Ito S, Verthelyi D. 2006. CpG oligodeoxynucleotides protect newborn mice from a lethal challenge with the neurotropic Tacaribe arenavirus. J Immunol 176:4940–4949. doi: 10.4049/jimmunol.176.8.4940. [DOI] [PubMed] [Google Scholar]
- 84.Gowen BB, Wong MH, Larson D, Ye W, Jung KH, Sefing EJ, Skirpstunas R, Smee DF, Morrey JD, Schneller SW. 2010. Development of a new tacaribe arenavirus infection model and its use to explore antiviral activity of a novel aristeromycin analog. PLoS One 5:e12760. doi: 10.1371/journal.pone.0012760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pythoud C, Rothenberger S, Martinez-Sobrido L, de la Torre JC, Kunz S. 2015. Lymphocytic choriomeningitis virus differentially affects the virus-induced type I interferon response and mitochondrial apoptosis mediated by RIG-I/MAVS. J Virol 89:6240–6250. doi: 10.1128/JVI.00610-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wolff S, Becker S, Groseth A. 2013. Cleavage of the Junin virus nucleoprotein serves a decoy function to inhibit the induction of apoptosis during infection. J Virol 87:224–233. doi: 10.1128/JVI.01929-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Garaigorta U, Chisari FV. 2009. Hepatitis C virus blocks interferon effector function by inducing protein kinase R phosphorylation. Cell Host Microbe 6:513–522. doi: 10.1016/j.chom.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sanchez A, Pifat DY, Kenyon RH, Peters CJ, McCormick JB, Kiley MP. 1989. Junin virus monoclonal antibodies: characterization and cross-reactivity with other arenaviruses. J Gen Virol 70:1125–1132. doi: 10.1099/0022-1317-70-5-1125. [DOI] [PubMed] [Google Scholar]
- 89.Buchmeier MJ, Lewicki HA, Tomori O, Oldstone MB. 1981. Monoclonal antibodies to lymphocytic choriomeningitis and Pichinde viruses: generation, characterization, and cross-reactivity with other arenaviruses. Virology 113:73–85. doi: 10.1016/0042-6822(81)90137-9. [DOI] [PubMed] [Google Scholar]
- 90.Stojdl DF, Lichty BD, tenOever BR, Paterson JM, Power AT, Knowles S, Marius R, Reynard J, Poliquin L, Atkins H, Brown EG, Durbin RK, Durbin JE, Hiscott J, Bell JC. 2003. VSV strains with defects in their ability to shutdown innate immunity are potent systemic anti-cancer agents. Cancer Cell 4:263–275. doi: 10.1016/S1535-6108(03)00241-1. [DOI] [PubMed] [Google Scholar]
- 91.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]