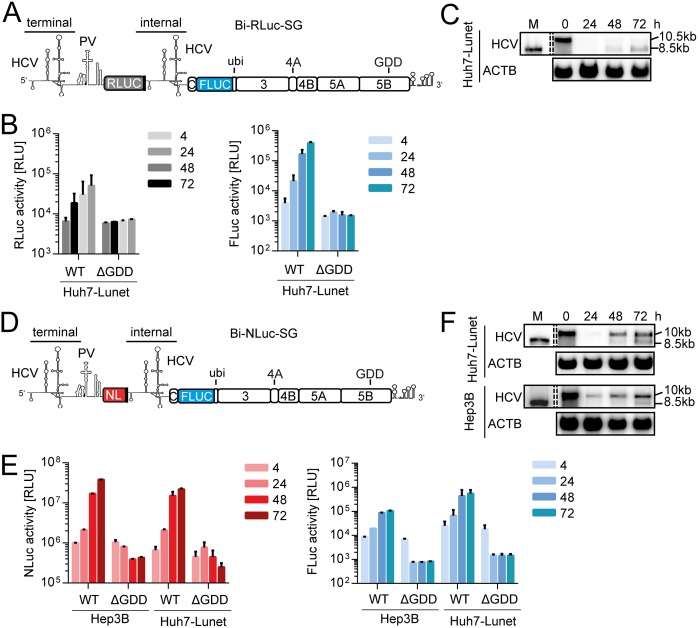
FIG 1.
Loss of full-length bicistronic replicon over time. (A) Schematic representation of Bi-RLuc-SG (positive strand). The bicistronic replicon features two HCV 5′-UTR regions. The terminal UTR is connected by a polioviral IRES (PV), governing translation of a Renilla luciferase reporter gene (RLuc, gray). After a short spacer element, containing several stop codons, the second cistron features the internal UTR, a firefly luciferase (FLuc) gene, fused to the HCV nonstructural proteins, and the 3′ UTR via a ubiquitin linker. (B) Luciferase assay for Renilla (gray) and firefly (blue) activity at 4 to 72 h postelectroporation using Bi-RLuc-SG WT and ΔGDD in Hep3B cells. (C) Corresponding Northern blot analysis of Bi-RLuc-SG levels, using a probe against HCV positive strand RNA (HCV) or β-actin (ACTB). Lane M, In vitro transcript of monocistronic replicon as a size marker (the lane was cropped from the same gel). 10.5 or 10 kb, size of the full-length replicon; 8.5 kb, size of the truncated variant. (D) Schematic representation of Bi-NLuc-SG. All features are as described in panel A, with the RLuc replaced by a Nano luciferase reporter (NL, red). (E) Luciferase assay of Nano (red) and Firefly (blue) activity 4 to 72 h posttransfection in Hep3B and Huh7-Lunet cells. (F) Corresponding Northern blot analysis of Bi-NLuc-SG RNA levels in both cell lines. ΔGDD, replication deficient NS5B mutant. Bars for luciferase activity represent the relative light units (RLU) without normalization (means ± the SD) of three biological replicates, each measured in technical duplicates. Note that transfections in Hep3B cells were supplemented with 50 pmol of miR-122.