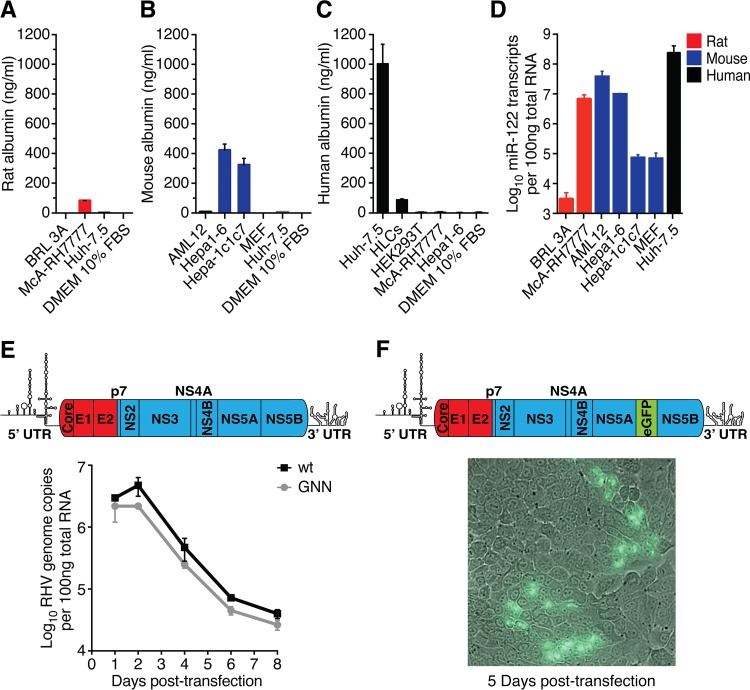
FIG 1.
Rodent liver-derived cell lines do not readily support productive RHV infection. (A to D) Quantification of liver-specific markers in hepatic cell lines. Secreted rat (A), mouse (B), and human (C) albumin was measured in cell culture supernatant. (D) Quantification of miR-122 in relevant cells. HLCs, hepatocyte-like cells derived from human induced pluripotent stem cells; MEF, mouse embryonic fibroblasts (negative control). Data points represent means ± standard deviations (SD) from duplicates. (E) Intracellular viral RNA after transfection of McA-RH7777 cells with full-length genomic RHV-rn1 RNA or the replication-deficient GNN control. Data points represent means ± SD from duplicates. Schematic of the full-length RHV genome depicting predicted protein coding regions and UTR structures is shown above (ORF drawn to scale; genes encoding structural [red] and nonstructural [blue] proteins are indicated). (F) Fluorescence microscopy after transfection of McA-RH7777 cells with full-length genomic RHV-rn1-eGFP RNA. A nonrepresentative picture taken 5 days posttransfection at ×40 magnification shows expression of eGFP in a few clusters of cells. Viral spread could not be detected. A genome schematic of the RHV-rn1-eGFP reporter virus is shown above.