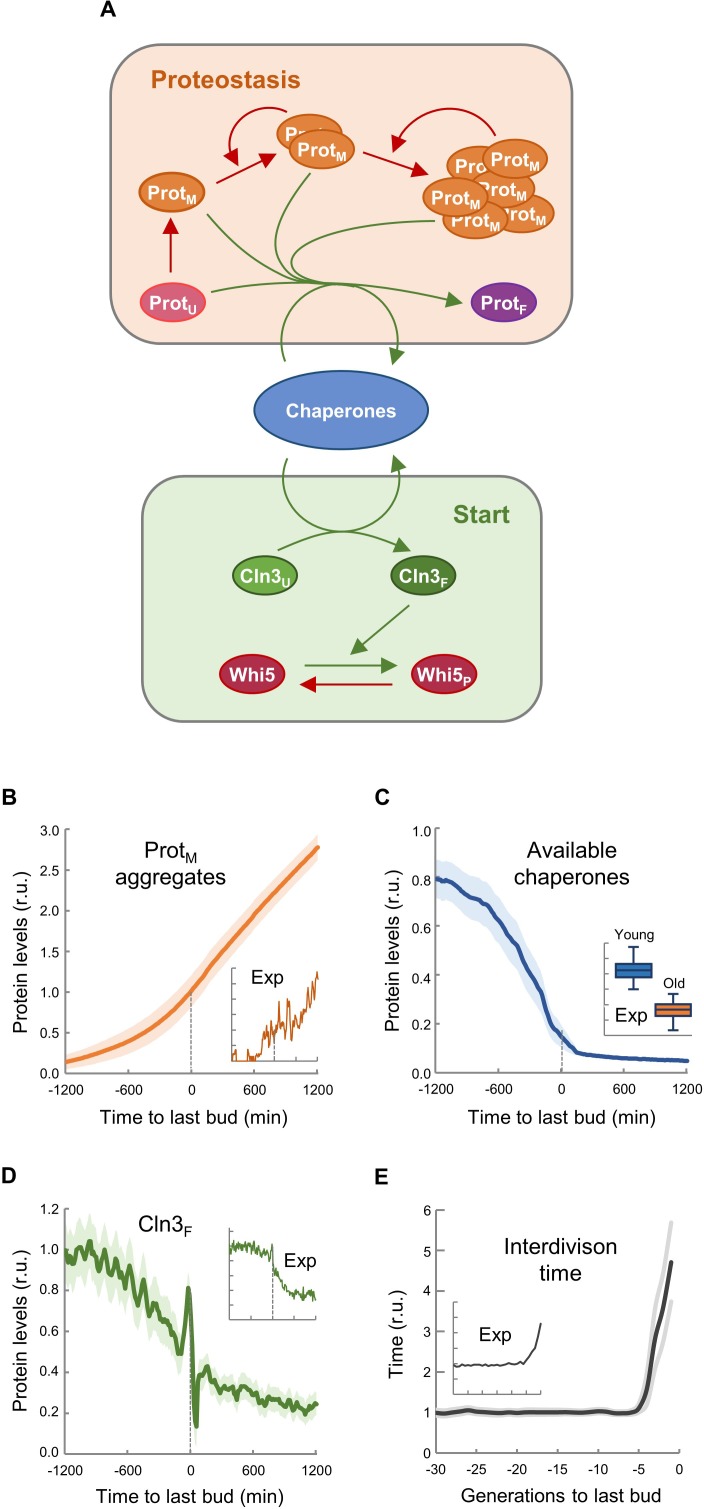
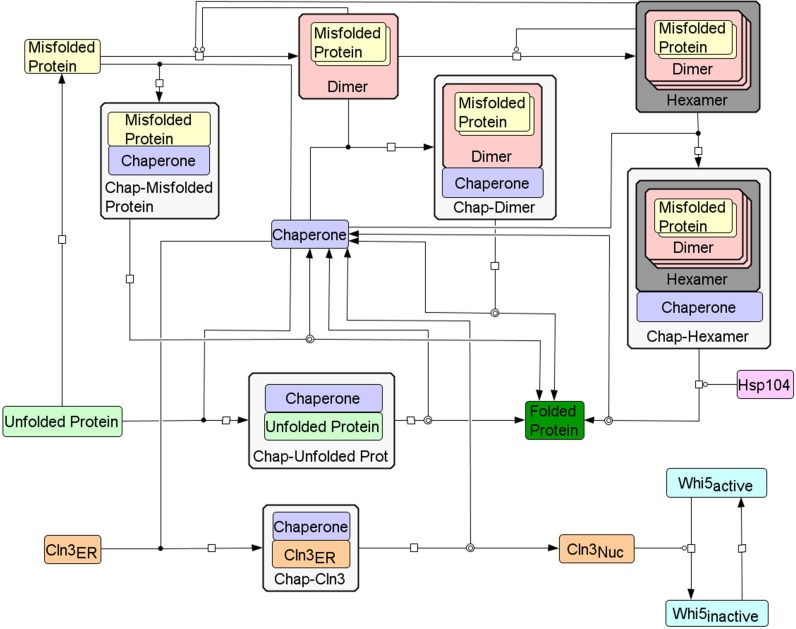
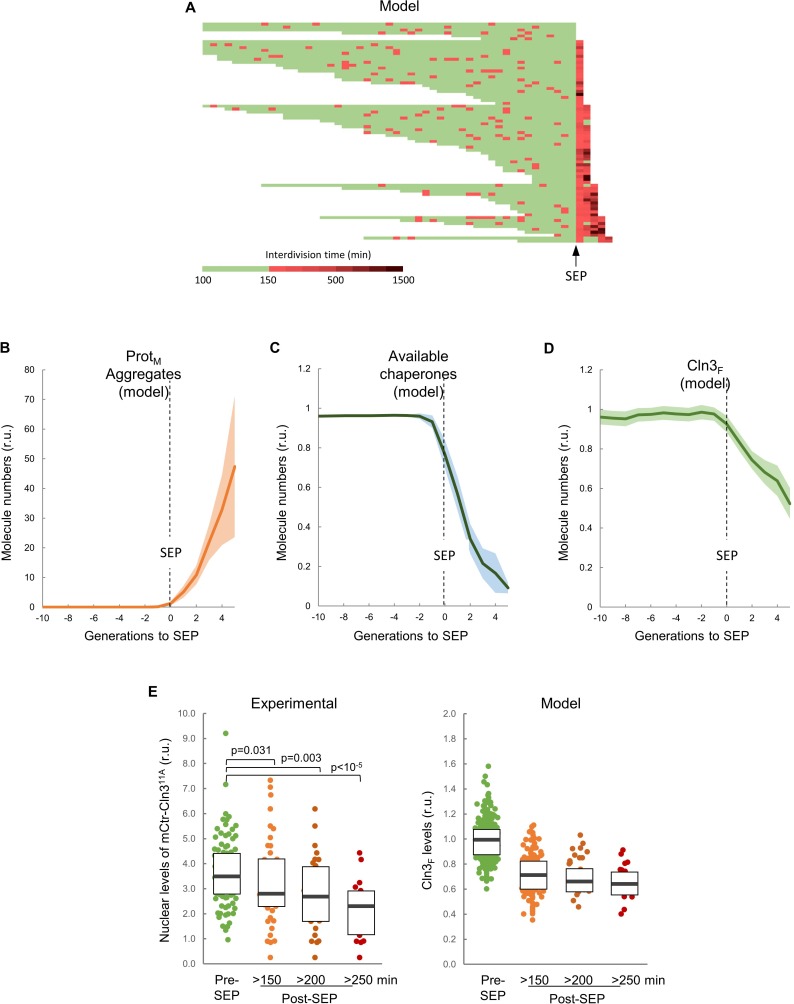
Figure 4. Asymmetric aggregate inheritance predicts a decrease in chaperone availability and a G1 arrest in aging cells.
(A) Scheme of the integrative mathematical model with chaperones playing concurrent roles in proteostasis and Start. (B–E) Predicted aggregate protein (B), available chaperone (C), and free folded Cln3 (D) levels and interdivision times in aging cells aligned at the last budding event. Values (mean ±CL, n = 75) are plotted as lines. Experimental (Exp) data from Figures 1B, H, 2F and 3E are also shown as insets for direct comparison.