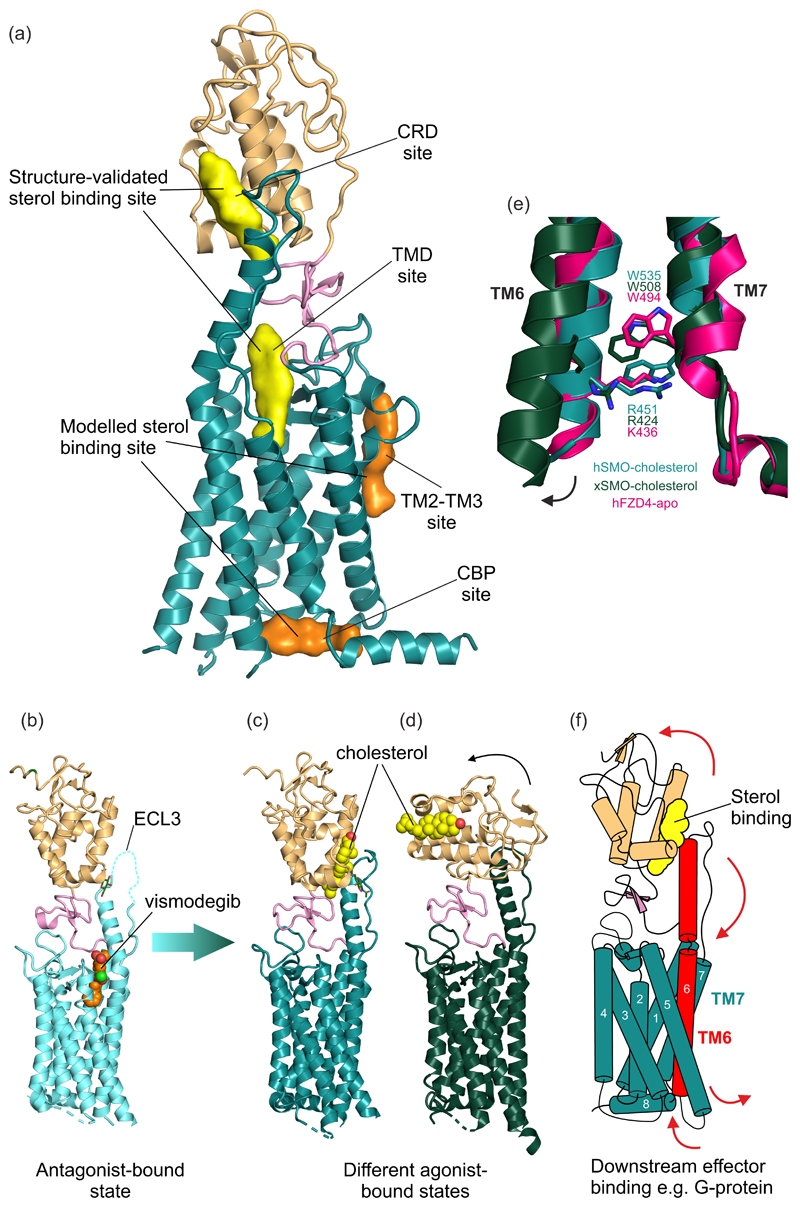
Figure 3. Structure of the HH signal transducer Smoothened.
(a) Multiple sterol binding sites have been proposed in SMO. In biochemical assays and crystal structures, cholesterol and oxysterols have been shown to bind to the CRD and the plant sterol-like molecule cyclopamine has been shown to bind in the TMD (shown in yellow). Two additional TMD sites (shown in orange) have been implicated using computational methods. (b) The structure of inhibited human SMO (hSMO) in complex with the antagonist vismodegib (PDB 5L7I [15]). Extracellular loop 3 (ECL3) is partially disordered (dashed line). Two potential active-state structures of SMO in complex with the agonist cholesterol are shown in (c) and (d). (c) The hSMO-cholesterol complex structure (PDB 5L7D [15]) shows SMO in an agonist-bound state, with the ordered ECL3 forming contacts with the CRD. (d) Xenopus SMO (xSMO)-cholesterol structure (PDB 6D35 [50]) reveals a more dramatic rotation of the CRD compared to the hSMO structure in (c), likely facilitated by removal of native glycans to facilitate crystallization. In (d) the TM6 moves outwards, as seen for other “canonical” GPCRs. (e) The cation-π lock is formed between a conserved tryptophan and basic residue at the cytoplasmic end of TM6 and 7 in class F GPCRs. hSMO-cholesterol (teal) shows an intact lock, while xSMO-cholesterol (dark green) shows the ruptured lock, perhaps indicative of activation. The lock is also seen in apo-Frizzled (PDB 6BD4 [61], hot pink). (f) Proposed model of transmembrane signalling by SMO. Sterol binding to the CRD causes its rotation relative to the TMD, causing a conformational change that is transmitted from the helical extension in ECL3 to TM6. This induces outward movement of TM6, rupturing the cation-π lock and exposing a new cytoplasmic surface for interaction with downstream effectors such as G-proteins.