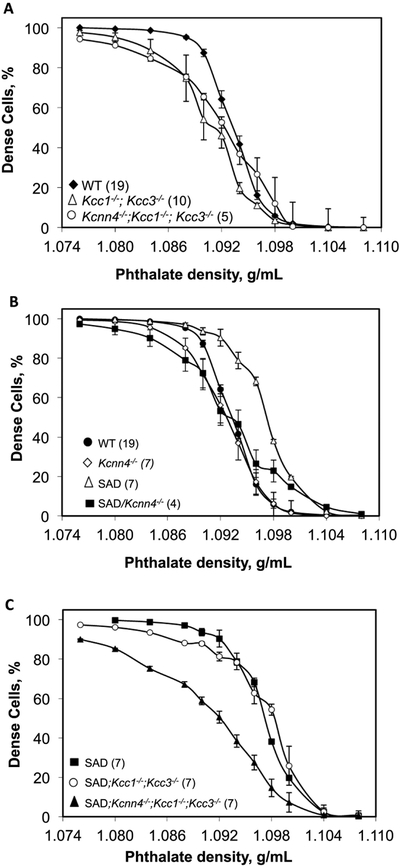
Fig. 3. Red cell phthalate density profiles as function of genotype.
A. Red cells from WT mice (black diamonds) compared with red cells from Kcc1−/−;Kcc3−/− mice (2xKO, open triangles) and red cells from Kcnn4−/−;Kcc1−/−;Kcc3−/− mice (3xKO, open circles). B. Red cells from WT (black circles, reproduced from data of panel A) and SAD mice (open triangles) compared with Kcnn4−/− mice (open diamonds) and SAD;Kcnn4−/− mice (black squares). WT mice in panels A and B included WTKcnn4 (n=8) and WTSAD (n=11), which were indistinguishable (not shown). C. Red cells from SAD mice (black squares, data replicated from panel B) compared with red cells from SAD;Kcc1−/−;Kcc3−/− mice (open circles) and red cells from SAD;Kcnn4−/−;Kcc1−/−;Kcc3−/− mice (black triangles). Values are means ± SEM for (n) independently tested pooled samples from each indicated genotype.