Fig. 1.
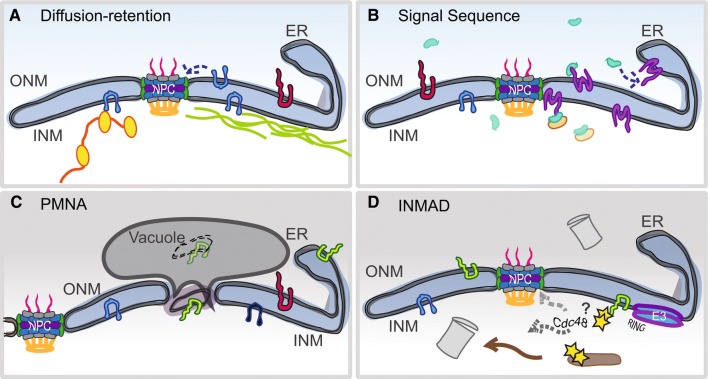
Post-translational mechanisms of INM homeostasis. a The diffusion retention model supports the idea that a NET (blue) is able to freely diffuse in and out of the nucleus as long as its cytoplasmic/nucleoplasmic domain is small enough to travel through the peripheral channel of NPCs (~ 60 kDa). NETs accumulate inside the nucleus by tethering to other nuclear factors such as chromatin (yellow) and/or lamins (green). b Using a sequence to signal a truncated version of karyopherin-α (green), an INM-bound NET (purple) is translocated through an NPC, followed by release of the karyopherin by association with Nup50/Nup2 (orange) or by other mechanisms. c Piecemeal nuclear autophagy (PMNA) occurs in the nuclei of yeast under nutrient deprivation, when sections of the INM are pinched off into the vacuole (yeast equivalent of the lysosome). PMNA is not known to occur in higher eukaryotes although there is evidence of autophagy of NE proteins (Dou et al. 2015). d Similar to ERAD, the INMAD pathway involves the ubiquitination of targets (yellow) by an E3 ligase containing a RING domain (blue–purple). Degradation of soluble targets may occur inside the nucleus by nuclear-localized proteasomes (gray). For membrane-bound targets, the Cdc48 AAA-ATPase may be involved in removal of the protein from the INM for degradation. Alternatively, retrotranslocation of the targets back to the ER for removal and processing by cytoplasmic ERAD machinery may occur