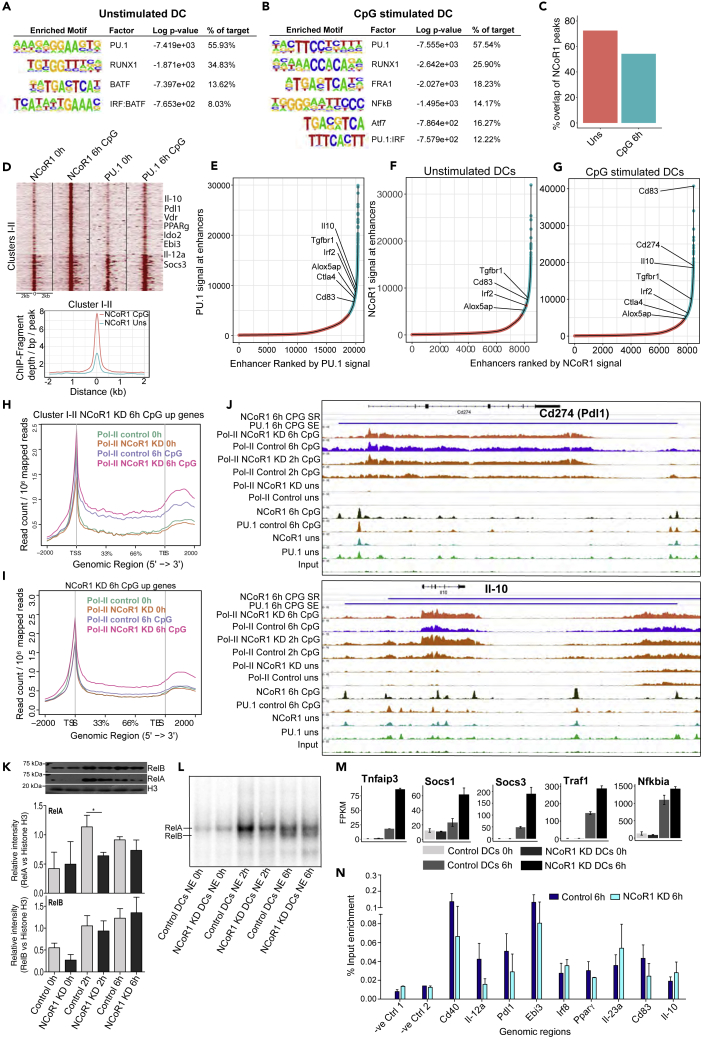
Figure 6.
NCoR1 Strongly Represses PU.1-Bound Super-Enhancers Present on Tolerogenic Genes after CpG Activation in CD8α+ cDCs
(A) Top significantly enriched de novo predicted DNA motifs of TFs in NCoR1 ChIP-seq peaks of unstimulated CD8α+ cDC1 DC line.
(B) Top significantly enriched DNA motifs of TFs in NCoR1 ChIP-seq peaks of 6-h CpG-stimulated CD8α+ cDC1 DCs.
(C) Bar graph showing the percentage overlap of NCoR1 with PU.1 ChIP-seq peaks before and after 6 h CpG activation.
(D) SeqMINER clustering showing clusters I-II depicting increased NCoR1 binding after CpG activation in cDC1 DC line when compared with unstimulated cells. Corresponding PU.1 ChIP-seq data lanes show its overlap with NCoR1-bound regions. Corresponding density plot shows the differential enrichment of reads in CpG compared with unstimulated at clusters I-II genomic regions. See also Table S6.
(E) Distribution of PU.1 ChIP-seq signals across the total PU.1-bound regulatory regions in CpG-activated CD8α+ DCs. Bound regions were ranked by the tag counts within the PU.1-enriched peaks to identify the super-enhancer regulatory regions or genes. Tolerogenic genes enriched in the PU.1 super-enhancer regions are marked. See also Table S7.
(F and G) Distribution of NCoR1 ChIP-seq signals across the total NCoR1-bound regulatory regions in unstimulated and CpG-activated CD8α+ DCs. Bound regions were ranked by the tag counts within the NCoR1-enriched peaks to identify the regulatory regions or genes super-repressed by NCoR1. Tolerogenic genes enriched in the NCoR1 super-repressed regions are marked. See also Table S7.
(H) Metagene plot showing the RNA Pol-II enrichment in unstimulated and 6-h CpG-stimulated control and NCoR1 KD DCs within the gene-body regions of the genes that are annotated to NCoR1 bound cluster I-II genomic regions.
(I) Metagene plot depicting the RNA Pol-II enrichment profile in unstimulated and 6 h CpG-stimulated control and NCoR1 KD DCs within the gene-body regions of the genes that are upregulated in 6 h CpG-activated NCoR1 KD DCs in RNA-seq data.
(J) IGV snapshots showing the ChIP-seq enrichments of NCoR1, PU.1, and RNA Pol-II at tolerogenic genes Il-10 and Pdl1 at 0 h and 6 h CpG-stimulated CD8α+ cDC1 DC line. RNA-Pol-II enrichment in CpG-activated control and NCoR1 KD DCs demonstrate the real-time transcription of these representative tolerogenic genes. The horizontal bars above the plot show the PU.1-bound super-enhancer (SE) regions that are strongly repressed (SR) by NCoR1 after CpG activation.
(K) Representative western blot picture for NF-κB subunits RelA and RelB depicting their nuclear translocation at 0 h, 2 h, and 6 h after CpG activation in control and NCoR1 KD CD8α+ cDC1 line. Corresponding densitometric analysis bar plots shows the relative intensity of RelA and RelB from four biological replicates (n = 4).
(L) Electrophoretic mobility shift assay (EMSA) demonstrating the binding activity of RelA and RelB at 0 h, 2 h, and 6 h after CpG activation. RelA and RelB bands are indicated by the marks.
(M) Fragment counts/kb/million read counts plots from control and NCoR1 KD RNA-seq data showing the differential expression of NF-κB negative regulators of NF-κB canonical signaling.
(N) RelA ChIP-qPCR depicting the enrichment of RelA at 10 randomly selected genomic regions when compared with negative control genomic regions.
p values are calculated using two-tailed paired Student's t test; fold change error is depicted as SEM. *p ≤ 0.05. See also Figures S6 and S7.