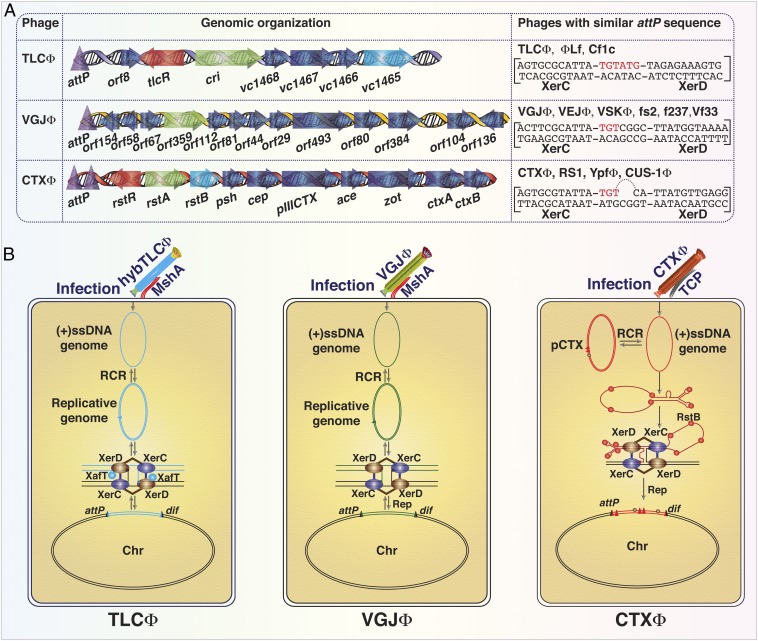
Fig. 1.
Insights into the IMEXs for their integration into chromosome dimer resolution site (dif). (A) Schematic diagram showing attachment sequences (attP) and the genomic organization of IMEXs associated with clinical and environmental isolates of V. cholerae. IMEXs are classified into 3 distinct classes based on the sequence and structure of their attP. Bases at the cross-over (CO) region that are important for strand exchanges are labeled in red. XerC and XerD binding sequences in the attP site of TLCΦ, VJGΦ, and CTXΦ are indicated within the parentheses. Arrows indicate different open reading frames (ORFs). Orientations of arrows specify transcription direction. Green arrows indicate ORF essential for phage replication. Red arrows indicate functions important for transcription regulation of phage genes. Sky blue arrows depict functions that are important for phage integration. (B) Schematic representation of the integration mechanisms of 3 classes of IMEXs. Each class of IMEX harbors distinct attachment sequences and follows a distinct mode of integration. While integration of IMEXs belonging to CTXΦ class is irreversible, integration of IMEXs belonging to the other 2 classes is reversible. The single-stranded genome (ssDNA) of TLCΦ used morphogenesis proteins of helper phage like fs2 and infected mannose-sensitive hemagglutinin (MshA) pili-positive V. cholerae. In the host cytoplasm, the ssDNA genome of TLCΦ converted into dsDNA and produced ssDNA phage genome by rolling circle replication (RCR). XerC and XerD recombinases recognize 28-bp attPTLC site of TLCΦ and mediate its integration with the help of XerD activating factor XafT. The microhomology at the overlap region between attPTLC and dif sites determines integration compatibility and strand exchanges. XerD mediates the first pair of strand exchanges and generates a Holliday junction. After isomerization, XerC mediates the second pair of strand exchanges and enables TLCΦ integration. TLCΦ integration is reversible, and excision probably follows the same sequences of strand exchanges as described for integration. The attP sequence in the dsDNA replicative form of V. cholerae Gillermo Javier filamentous phage (VGJΦ) harbors dif like XerC- and XerD-binding sites separated by a 7-bp CO region. Three base pairs of the CO region immediately adjacent to the XerC-binding site are identical to those of the dif1 sequence. The pseudo-Holliday junction resulting from the strand exchanges between attPVGJ and dif1 catalyzed by XerC is converted into product by chromosomal replication. The cholera toxin encoding CTXΦ recognizes host-encoded toxin coregulated pilus (TCP) as receptor and introduce its (+)ssDNA genome into host cytoplasm. The (+)ssDNA genome CTXΦ is either converted to replicative dsDNA or directly integrated into the host chromosome. For integration, the ∼150-nucleotide-long attachment site folds into a fork hairpin structure where inversely oriented attP1 and attP2 form intrastrand base pairing and generates functional attP(+) site. XerC mediates 1 pair of strand exchanges and generates a pseudo-Holliday junction. Host DNA replication resolves the pseudo-Holliday junction and completes the irreversible integration of CTXΦ.