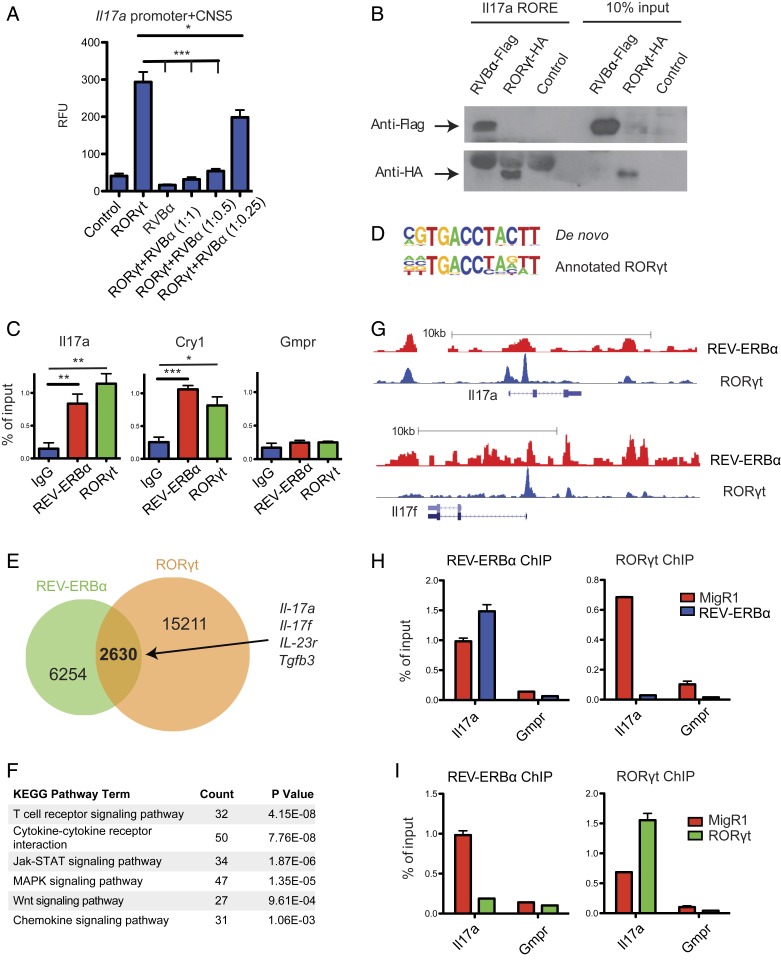
Fig. 2.
REV-ERBα directly competes with RORγt and represses Th17 signature gene expression. (A) Luciferase assays of EL4 T cells cotransfected with an Il17a luciferase reporter, and combinations of RORγt and REV-ERBα at various ratios, with the amount of RORγt transfected remaining constant. Renilla luciferase activity was used as internal control. (B) Western blot to detect the binding of HA-tagged RORγt and Flag-tagged REV-ERBα expressed in CD4 T cells to biotinylated oligonucleotides containing RORE motif derived from the Il17a CNS5 enhancer. (C) ChIP-qPCR to detect the binding of Il17a CNS5 enhancer, Cry1 (positive control) and Gmpr (negative control) by REV-ERBα and RORγt in Th17 cells. (D–G) Analysis of Th17 cell REV-ERBα ChIP-seq data along with published RORγt ChIP-seq data. (D) Alignment of de novo generated REV-ERBα binding sequence to annotated RORγt binding sequence. (E) Venn diagram depicting the numbers of unique and shared genes bound by REV-ERBα and RORγt. (F) KEGG pathway analysis of REV-ERBα bound genes. (G) Trace analysis of ChIP-seq data visualized on the UCSC genome browser showing overlapping binding sites of REV-ERBα and RORγt at Il17a and Il17f loci. (H and I) ChIP-qPCR to detect changes in REV-ERBα and RORγt binding to the Il17a locus in response to ectopic expression of REV-ERBα (H) or RORγt (I). Data represents mean ± SEM. Statistical analyses were performed using unpaired 2-tailed Student’s t test (*P < 0.05, **P < 0.01, ***P < 0.001).