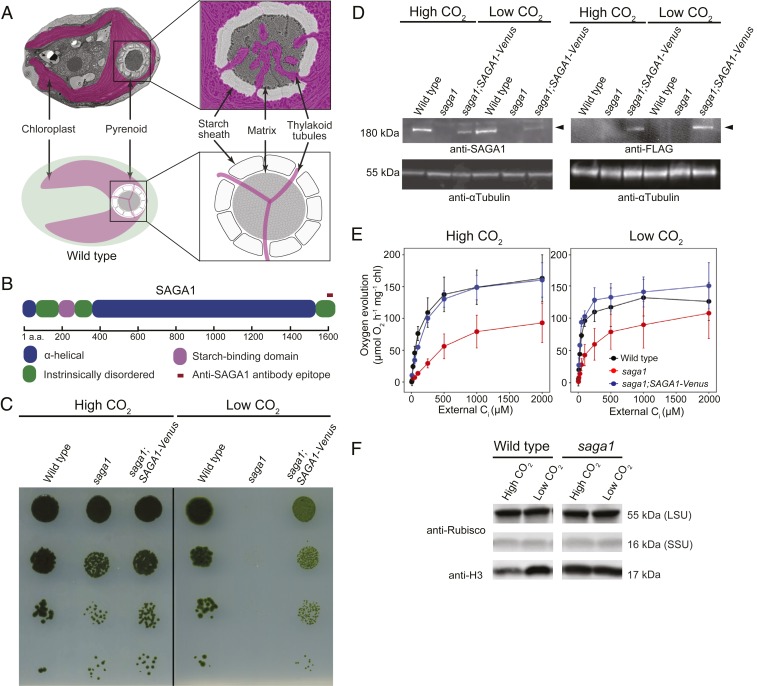
Fig. 1.
SAGA1, a gene with a predicted starch-binding domain and alpha helical stretch, is necessary for a functional CCM. (A) A Chlamydomonas cell is shown with the substructures of the pyrenoid labeled. (B) SAGA1 has a predicted starch binding domain followed by a long predicted alpha helical region. (C) Agar growth phenotypes of wild type, saga1, and saga1;SAGA1-Venus are shown. Serial 1:10 dilutions were spotted on TP minimal medium and grown at high (4%) and low (0.04%) CO2 under 500 μmol photons m−2⋅s−1 illumination. (D) SAGA1 protein levels in wild-type, saga1, and saga1;SAGA1-Venus cells grown at low and high CO2 were probed with an anti-SAGA1 polyclonal antibody and with an anti-FLAG antibody. Anti-tubulin is shown as a loading control. (E) Oxygen evolution is shown as a function of external Ci for cells grown in high and low CO2. (F) Rubisco protein levels in wild type and saga1 grown at low and high CO2 were probed with a polyclonal antibody raised to Rubisco. Anti-histone H3 is shown as a loading control. LSU: large subunit; SSU: small subunit.