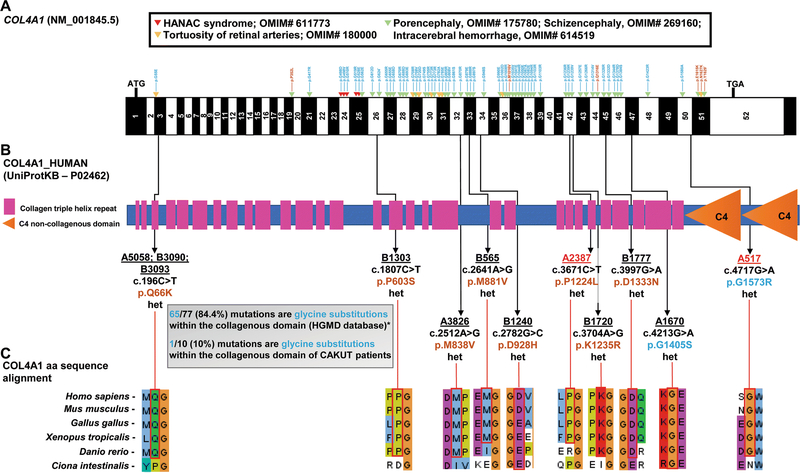
Figure 1. WES detects in 12 out of 550 CAKUT families with isolated CAKUT heterozygous COL4A1 mutations, that are mostly non-glycine substitutions, as opposed to the established COL4A1-related disorders that mostly report glycine substitutions.
A) Exon structure of human COL4A1 cDNA is denoted in black and white; ATG, start codon; TGA, stop codon; arrow heads denote missense mutations in COL4A1-related disorders found in the HGMD® database: blue denotes glycine substitutions, while brown denotes non-glycine substitutions. Note that 65 out of 77 (84.4%) mutations are glycine substitutions.
*Only 64 of the 77 mutations of the HGMD® database are depicted due to space limitations.
B) Protein domain structure of human COL4A1 showing the domain position of each of the missense mutations identified in ten families with CAKUT. Family ID is underlined; family ID is highlighted in red if extra-renal clinical manifestations are noted, p.change is in brown for non-glycine substitutions and in blue for glycine substitutions. Note that only one out of ten mutations (10%) is a glycine substitution within the collagenous domain.
C) Clustal alignment of amino acid sequences of COL4A1 demonstrating evolutionary conservation from mammalia to ascidiacea for each amino acid residue.