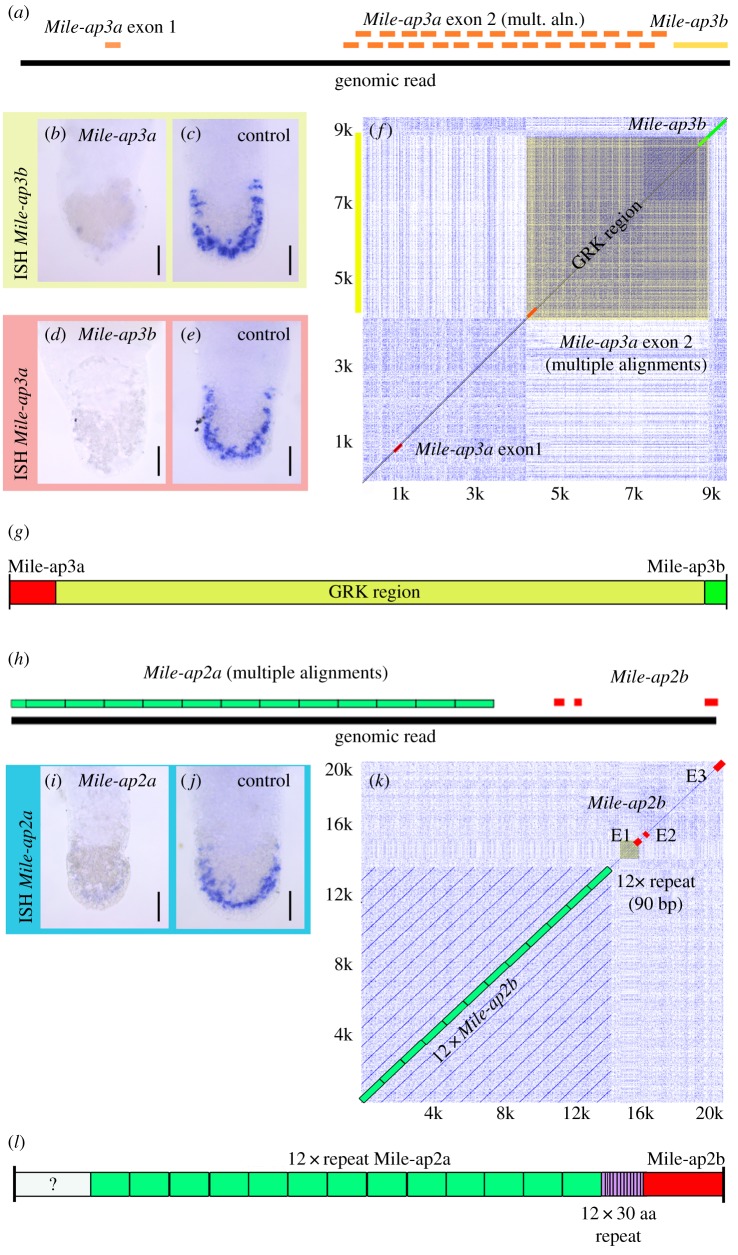
Figure 5.
Oxford Nanopore sequencing of gDNA. (a) Mapping of Mile-ap3a and Mile-ap3b onto a single Oxford Nanopore read. (b,c) RNAi knock-down of Mile-ap3a and in situ hybridization with a probe of Mile-ap3b. mult. aln., multiple alignments. (d,e) RNAi knock-down of Mile-ap3b and in situ hybridization with a probe of Mile-ap3a. (f) Dot plot of a nine kb section of genomic read ‘27752bc6-c8c2-433f-8100-0d294e375058’. (g) Hypothetical adhesive protein containing Mile-ap3a and Mile-ap3b and an extensive GRK low complexity region. (h) Mapping of Mile-ap2a and Mile-ap2b onto a single Oxford Nanopore read. (i,j) RNAi knock-down of Mile-ap2a and in situ hybridization with a probe of Mile-ap2b. (k) Dot plot of a 20 kb section of genomic read ‘7de1b9a2-7996-4a1a-8b10-3f661449fd01’. (l) Hypothetical adhesive protein containing Mile-ap2a and Mile-ap2b and 12 repeats of Mile-ap2a. ISH, in situ hybridization.