Figure EV4. The interactome of C/EBPα reveals a preferential interaction with SWI/SNF, NURD, KDM1A/HDAC/REST, and RSF chromatin remodeling complexes.
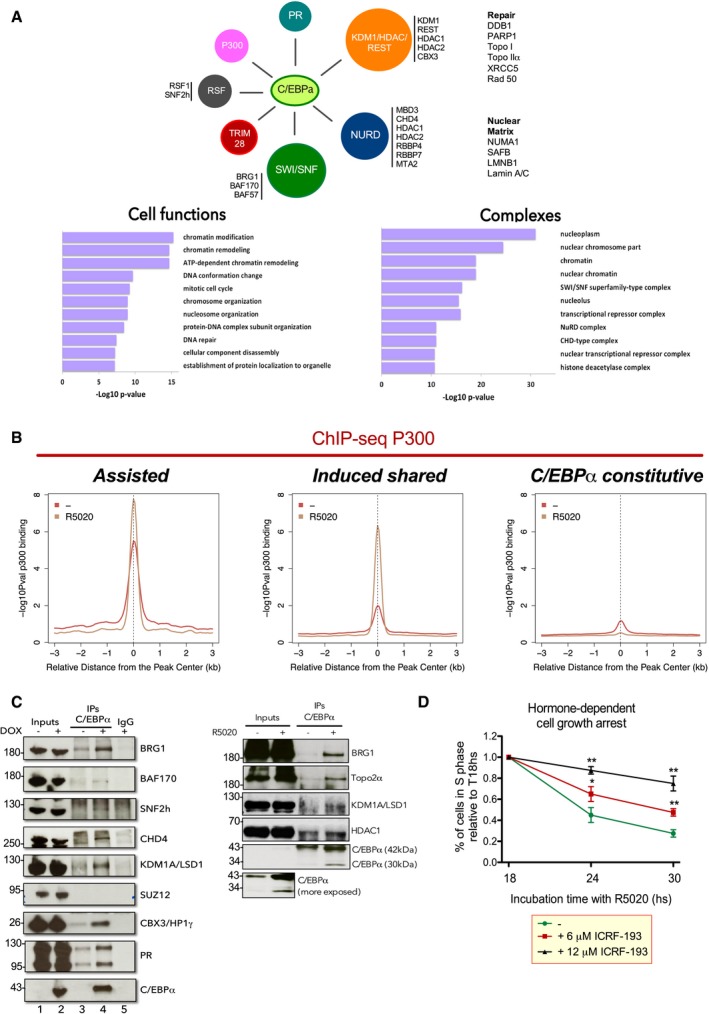
- Immunoprecipitation of C/EBPα followed by mass spec in a T47D cell line expressing inducible C/EBPα (see Materials and Methods). Lower panels: Analysis of the peptides showed that the interacting proteins function in chromatin remodeling, chromatin modification, mitotic cell cycle, DNA conformation change, DNA repair, and nucleosome organization. Right panel: By using the CORUM database (Ruepp et al, 2008), we identify SWI/SNF (BAF), NURD, and CHD complexes as well as histone deacetylases.
- Analysis of the ChIP‐seq of P300 in T47D cells treated or not with hormone in “assisted”, “induced shared”, “PR‐exclusive”, “C/EBPα‐constitutive”, and “C/EBPα‐exclusive” regions.
- Validation of the C/EBPα interactors by co‐IP in T47DindC/EBPα cells −/+ dox (left panel) and T47D cells −/+ hormone (right panel). The immunoprecipitates (IP) were analyzed by immunoblotting with specific antibodies for C/EBPα, Topo 2α, BRG1, BAF170, SNF2h, CHD4, KDM1, CBX3, and PR.
- T47D cells treated or not with 6 and 12 μM of ICRF193 and 10 nM R5020 as indicated were subjected to flow cytometric analysis of cell cycle to quantify the cells in S phase. Each value corresponds to the mean ± SD of three experiments performed in duplicate. **P < 0.01, *P < 0.05 using Student's t‐test.
Source data are available online for this figure.
