Figure EV4. Overproduction of TagA inhibits T6SS in V. cholerae .
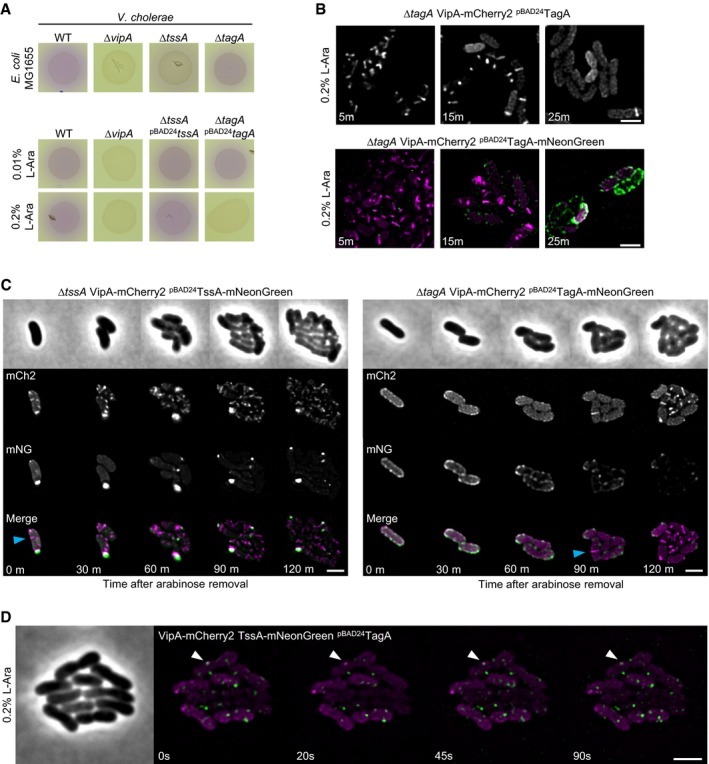
- Bacterial competition assay. Killing of E. coli (MG1655, lacZ+) by V. cholerae T6SS can be observed as CPRG substrate is converted by released β‐galactosidase over time (red color). ΔtssA VC strain shows significantly decreased killing efficiency, while tagA VC knockout is indistinguishable from WT killing efficiency. ΔvipA strain was used as T6SS‐negative control (upper panel). Overproduction of TagAVC but not TssAVC leads to inhibition of T6SS‐mediated killing (lower panel).
- Overproduction of TagAVC and TagAVC‐mNeonGreen leads to inhibition of T6SS sheath formation. Overproduced TagAVC‐mNeonGreen localized predominantly at the cell periphery in discrete foci. Scale bars: 2 μm.
- Indicated strains were pre‐induced with 0.2% l‐arabinose for 1 h, then washed, and observed using fluorescence microscopy. Dynamic structures (blue arrow) can be detected directly after washing in the TssAVC‐mNeonGreen overproducing strain (left panel). The strain overproducing TagAVC‐mNeonGreen was only able to form T6SS structures after 90‐min recovery period (right panel). Scale bar: 2 μm.
- Time lapse series of a VipA‐mCherry2 TssAVC‐mNeonGreen double‐tagged strain overproducing TagAVC from plasmid. TagAVC expression was induced for 1 h with 0.2% l‐arabinose prior to imaging. TssAVC‐mNeonGreen displays non‐dynamic behavior and co‐localizes with sheath foci in the cell periphery (white arrow). Scale bar: 2 μm.
