Acute lymphoblastic leukemia (ALL) is the most common form of pediatric malignancy, encompassing multiple entities characterized by both distinct clinical and biological features as well as treatment response.1 Underlying genetic alterations is a key determinant to explain such heterogeneity and inform disease classification, especially among B-cell precursor (BCP)-ALL.2 Thus, high hyperdiploidy and the ETV6-RUNX1 fusion are associated with a favorable outcome. By contrast, KMT2A rearrangements and the BCR-ABL1 fusion confer adverse prognosis. Until recently, a large proportion of BCP-ALL patients lacking such aberrations (named B-other ALL) was allocated to the intermediate prognostic subgroup.3 Great advances have been made to unravel the genetics of this subgroups leading to the discovery of an entity of high-risk ALL with BCR-ABL1-like properties.4 Patients with BCR-ABL1-like ALL have a gene expression profile similar to that of BCR-ABL1-positive ALL but lack the BCR-ABL1 fusion gene. BCR-ABL1-like ALL has a poor outcome and frequently harbors deletions in IKZF1 or other B-lymphoid key genes.4 Detection of BCR-ABL1-like ALL is clinically important due to the presence of targetable kinase-activating alterations, which represent the hallmark of this entity.5 Here, we report the case of a young adult with a refractory BCP-ALL displaying a rare ZC3HAV1-ABL2 fusion. ABL2 is a member of the Abelson family of nonreceptor tyrosine kinase proteins closely related to ABL1. To date, the efficacy of tyrosine kinase inhibitors (TKI) on this particular genetic alteration remains unknown.5
The patient was an 18-year-old male, with no previous medical history, referred to our Hematology Department for hyperleukocytosis incidentally discovered during a blood donation. The full blood count showed: white blood cell count 171 × 109/L (including 90% of blast cells), hemoglobin concentration 9.3 g/dL, and platelet count 107 × 109/L. Clinical examination demonstrated asthenia, pallor, and splenomegaly without adenopathy. Bone marrow aspirate was of high cellularity with 90% of peroxydase negative-blast cells. Additionally, a lumbar puncture showed central nervous system involvement. Immunophenotyping confirmed the diagnosis of BCP-ALL with the expression of CD34, HLA-DR, and B lymphoid markers CD19, CD10, CD20, and CD22 and the negativity of myeloid and T-lymphoid markers. Conventional cytogenetics showed a normal karyotype 46,XY in all metaphases. Fluorescent in situ hybridization (FISH) and molecular studies were negative for ETV6-RUNX1, BCR-ABL1, TCF3-PBX1, and KMT2A fusions. IKZF1 deletion encompassing exons 4 to 8, as well as PAX5 and EBF1 deletions were detected by multiplex ligation probe amplification (MLPA P335 kit, MRC Holland, Amsterdam, the Netherlands).6
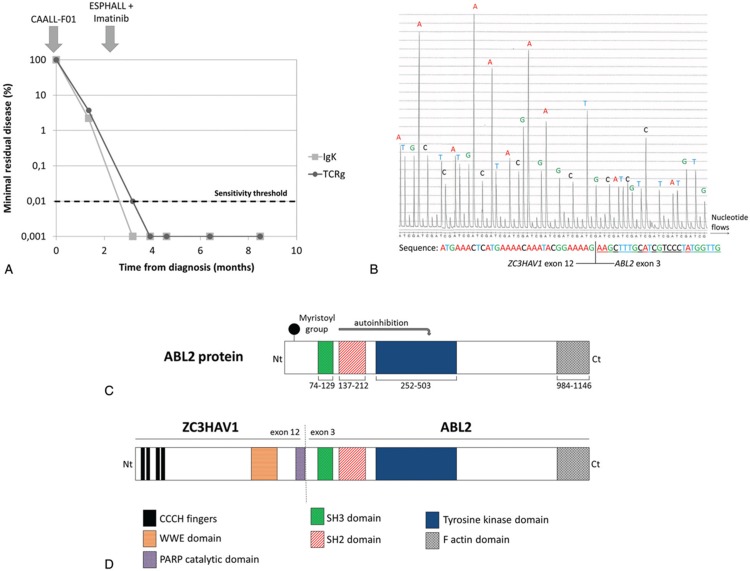
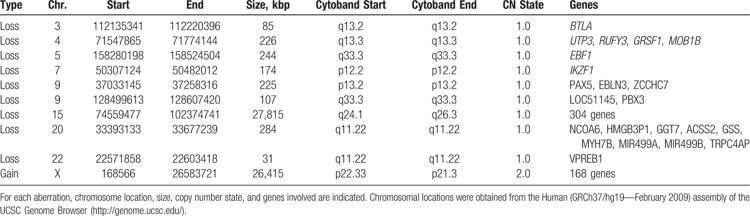
First-line treatment was initiated according to the CAALL-F01 clinical trial (A French Protocol for the Treatment of Acute Lymphoblastic Leukemia in Children and Adolescents; ClinicalTrials.gov Identifier NCT02716233). Evaluation after 7 days of prednisolone revealed the persistence of 6.9 × 109/L blast cells in the peripheral blood. The patient was then treated in the high-risk group. Induction therapy included vincristine 1.5 mg/m2 IV (D8, D15, D22, and D29), daunorubicin 30 mg/m2 IV (D8, D15, D22, and D29), prednisolone 60 mg/m2 per day PO (D1–D7, and D8–D28 then tapered over 1 week), peg-asparaginase 2500 IU/m2 IV (D12 and D26), intrathecal methotrexate (D1), and triple intrathecal (methotrexate, cytarabine, and corticoid; D13 and D24). Bone marrow aspirate at D35 revealed induction failure with positivity of minimal residual disease (MRD) above 10−2 for Ig/TCR markers using standard procedures7 (approximately 3% residual leukemic blasts; Fig. 1A) and for leukemia-associated immunophenotype in flow cytometry (not shown). Considering the patient to have a B-other ALL, together with age and poor treatment response, we performed reverse transcriptase multiplex ligation probe amplification (RT-MLPA) to extend the panel of transcript fusion detection8 as well as single nucleotide polymorphism (SNP)-array karyotyping. Remarkably, the RT-MLPA detected a fusion between the exon 12 of ZC3HAV1 and the exon 3 of ABL2 exon 3 (Fig. 1B). Additionally, SNP-array (Cytoscan High Density, Affymetrix, Thermo Fisher Scientific, Waltham, Massachusetts, United States) confirmed deletions in IKZF1 (7p12.2), EBF1 (5q.33.3), and PAX5 (9p13.2) and revealed small deletions in BTLA (3q12.2), PBX3 (9q33.3), VPREB1 (22q11.22), as well as larger copy-number aberrations at 15q and Xp regions (Table 1).
Figure 1.
Clinical and molecular data. (A) Minimal residual disease monitoring in bone marrow using rearrangements of immunoglobulin light chain kappa and T-cell receptor gamma as leukemia-specific markers. (B) ZC3HAV1-ABL2 fusion sequence determined by reverse transcriptase-multiplex ligation probe amplification. The sequencing is performed by cyclic flowing of nucleotides (A, T, C, G). Each nucleotide incorporation gives a strong signal (peak indicated with A, T, C or G), which is proportional to the number of nucleotides incorporated. The sequence deduced is shown below with indication of the ZC3HAV1 part and ABL2 parts. (C) Domain organization of ABL2. (D) Domain organization of the putative ZC3HAV1-ABL2 fusion protein.
Table 1.
Copy-Number Abnormalities Found by Single Nucleotide Polymorphism Array.
The ZC3HAV1-ABL2 fusion has been rarely described in BCP-ALL.9,10 The ZC3HAV1 gene (Zinc Finger CCCH-Type Containing, Antiviral 1), located at 7q34, encodes an RNA-binding protein, which acts as a tumor suppressor and regulates various processes during cell development and homeostasis.11 The ABL2 gene (Abelson tyrosine-protein kinase 2), located at 1q25.2, encodes a nonreceptor tyrosine kinase protein that belongs to the Abelson family. ABL2 shares a high degree of sequence conservation and a similar domain organization with ABL1. ABL2 functions may overlap with those of ABL1 and include cytoskeleton organization, cell proliferation, adhesion, and migration.12 Activity of ABL2 is regulated by an autoinhibitory mechanism. Notably, ABL2 is maintained in an inactive conformation by an N-terminal myristoyl group binding to a hydrophobic pocket in the kinase domain10 (Fig. 1C). Thus, the fusion of ZC3HAV1 to the N-terminal part of ABL2 is supposed to disrupt this autoinhibitory mechanism and enhance the tyrosine kinase activity to induce leukemia (Fig. 1D). Additionally, crystallization experiments of the ZC3HAV1 protein have revealed that its N-terminal part, containing the 4 tandem CCCH-type zinc-finger motifs, was involved in protein dimerization,13 a feature shared with many ABL1 fusion partners.
Considering those findings, our patient was subsequently treated according to the ESPHALL trial (ClinicalTrials.gov Identifier NCT00287105).14 He received imatinib 500 mg/d (D1–D21) in combination with dexamethasone, vincristine, high-dose methotrexate, high-dose cytarabine, high-dose asparaginase, and cyclophosphamide. He finally achieved complete remission after the first consolidation course with negativity of MRD based on Ig/TCR monitoring and immunophenotyping. Due to this complete response, the indication of hematopoietic stem cell transplantation was not retained. Subsequent MRD evaluations confirmed MRD negativity (Fig. 1A).
BCR-ABL1-like ALL is a common subtype of ALL, especially in adolescents and young adults (AYA) in whom it could account for more than 25% of BCP-ALL cases.15 Identifying such ALL is essential since patients could benefit of treatment combinations with TKIs. Particularly, significant results have been reported with TKIs in patients with chemotherapy-refractory ALL harboring ABL116 or PDGFRB17 fusions while experience in patients with ABL2 fusions is limited.10 This report suggests that ABL2 fusions could respond to TKIs in vivo. Further studies and international collaboration are required to better define management and prognosis of patients bearing these rare rearrangements. In clinical practice, the detection of BCR-ABL1-like fusions, which are diverse and often cryptic, can be assessed by FISH analyses (using split FISH probes targeting the tyrosine kinase genes) or molecular methods such as RT-MLPA or RNA sequencing.18 Because of major treatment implications, such experiments should be consistently performed in patients with B-other ALL and/or suggestive findings (AYA, poor treatment response, evocative pattern of microdeletions by MLPA or SNP-array).
Acknowledgment
The authors thank the Lille Hospital Tumor Bank (certification NF 96900-2014/65453-1), for handling, conditioning, and storing patient samples.
Footnotes
Citation: Decool G, Domenech C, Grardel N, Plesa A, Raczkiewicz I, Ducourneau B, Ruminy P, Pages M-P, Girard S, Fenwarth L, Preudhomme C, Bertrand Y, Duployez N. Efficacy of Tyrosine Kinase Inhibitor Therapy in a Chemotherapy-refractory B-cell Precursor Acute Lymphoblastic Leukemia With ZC3HAV1-ABL2 Fusion. HemaSphere, 2019;0:0. http://dx.doi.org/10.1097/HS9.0000000000000193
Funding/support: None.
Disclosure: The authors have indicated they have no potential conflicts of interest to disclose.
Authorship contributions: GD, ND, IR, BD, PR, LF, NG, and CP performed molecular analyses. AP performed flow cytometry. M-PP performed cytogenetic analyses. SG performed morphological diagnosis. CD and YB provided clinical data. GD, ND, CP, and YB wrote the manuscript which was approved by all coauthors.
References
- 1.Mullighan CG. The genomic landscape of acute lymphoblastic leukemia in children and young adults. Hematology Am Soc Hematol Educ Program. 2014;2014:174–180. [DOI] [PubMed] [Google Scholar]
- 2.Arber DA, Orazi A, Hasserjian R, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127:2391–2405. [DOI] [PubMed] [Google Scholar]
- 3.Schwab C, Harrison CJ. Advances in B-cell precursor acute lymphoblastic leukemia genomics. HemaSphere. 2018;2:e53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Den Boer ML, van Slegtenhorst M, De Menezes RX, et al. A subtype of childhood acute lymphoblastic leukaemia with poor treatment outcome: a genome-wide classification study. Lancet Oncol. 2009;10:125–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roberts KG, Morin RD, Zhang J, et al. Genetic alterations activating kinase and cytokine receptor signaling in high-risk acute lymphoblastic leukemia. Cancer Cell. 2012;22:153–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moorman AV, Enshaei A, Schwab C, et al. A novel integrated cytogenetic and genomic classification refines risk stratification in pediatric acute lymphoblastic leukemia. Blood. 2014;124:1434–1444. [DOI] [PubMed] [Google Scholar]
- 7.van der Velden VHJ, Cazzaniga G, Schrauder A, et al. Analysis of minimal residual disease by Ig/TCR gene rearrangements: guidelines for interpretation of real-time quantitative PCR data. Leukemia. 2007;21:604–611. [DOI] [PubMed] [Google Scholar]
- 8.Ruminy P, Marchand V, Buchbinder N, et al. Multiplexed targeted sequencing of recurrent fusion genes in acute leukaemia. Leukemia. 2016;30:757–760. [DOI] [PubMed] [Google Scholar]
- 9.Lilljebjörn H, Henningsson R, Hyrenius-Wittsten A, et al. Identification of ETV6-RUNX1-like and DUX4-rearranged subtypes in paediatric B-cell precursor acute lymphoblastic leukaemia. Nat Commun. 2016;7:11790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen P, Chu A, Zia H, et al. CD25 expression in B lymphoblastic leukemia/lymphoma predicts t(9;22)(q34;q11)/Philadelphia chromosome translocation (Ph) and is associated with residual disease in Ph-negative patients. Am J Clin Pathol. 2016;146:632–638. [DOI] [PubMed] [Google Scholar]
- 11.Todorova T, Bock FJ, Chang P. Poly(ADP-ribose) polymerase-13 and RNA regulation in immunity and cancer. Trends Mol Med. 2015;21:373–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salah E, Ugochukwu E, Barr AJ, et al. Crystal structures of ABL-related gene (ABL2) in complex with imatinib, tozasertib (VX-680), and a type I inhibitor of the triazole carbothioamide class. J Med Chem. 2011;54:2359–2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen S, Xu Y, Zhang K, et al. Structure of N-terminal domain of ZAP indicates how a zinc-finger protein recognizes complex RNA. Nat Struct Mol Biol. 2012;19:430–435. [DOI] [PubMed] [Google Scholar]
- 14.Biondi A, Gandemer V, De Lorenzo P, et al. Imatinib treatment of paediatric Philadelphia chromosome-positive acute lymphoblastic leukaemia (EsPhALL2010): a prospective, intergroup, open-label, single-arm clinical trial. Lancet Haematol. 2018;5:e641–e652. [DOI] [PubMed] [Google Scholar]
- 15.Roberts KG, Mullighan CG. Genomics in acute lymphoblastic leukaemia: insights and treatment implications. Nat Rev Clin Oncol. 2015;12:344–357. [DOI] [PubMed] [Google Scholar]
- 16.Duployez N, Grzych G, Ducourneau B, et al. NUP214-ABL1 fusion defines a rare subtype of B-cell precursor acute lymphoblastic leukemia that could benefit from tyrosine kinase inhibitors. Haematologica. 2016;101:e133–e134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lengline E, Beldjord K, Dombret H, et al. Successful tyrosine kinase inhibitor therapy in a refractory B-cell precursor acute lymphoblastic leukemia with EBF1-PDGFRB fusion. Haematologica. 2013;98:e146–e148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boer JM, den Boer ML. BCR-ABL1-like acute lymphoblastic leukaemia: from bench to bedside. Eur J Cancer. 2017;82:203–218. [DOI] [PubMed] [Google Scholar]