Supplemental Digital Content is available in the text
Abstract
T-cell Receptor Gamma (TRG) rearrangements are commonly used to detect clonal lymphoproliferations in hematopathology, since they are rearranged in virtually all T lymphocytes and have a relatively limited recombinatorial repertoire, which reduces the risk of false negative results, at the cost of potential false positivity. We developed an initial one-tube, 2-fluorochrome EuroClonality TRG PCR multiplex (TRG-1T-2F) which was compared to the original 2-tube, 2-fluorochrome EuroClonality/BIOMED-2 TRG PCR (TRG-2T-2F) and a commercial Invivoscribe one-tube, one-fluorochrome kit (IVS-1T-1F) on a series of 239 samples, including both T-cell malignancies and reactive cases. This initial assay yielded discrepant results between the 10 participating EuroClonality laboratories when using 2 fluorochromes, leading to adoption of a final single color EuroClonality strategy (TRG-1T-1F). Compared to TRG-2T-2F, both TRG-1T-1F and IVS-1T-1F demonstrated easier interpretation and a lower risk of false positive from minor peaks in dispersed repertoires. Both generate smaller fragments and as such are likely to be better adapted to analysis of formalin-fixed paraffin-embedded (FFPE) tissue samples. Their differential performance was mainly explained by (i) superposition of biallelic rearrangements with IVS-1T-1F, due to more extensive overlapping of the repertoires and (ii) intentional omission of the TRGJP primer in TRG-1T-1F, in order to avoid the potential risk of confusion of consensus TRG V9-JP normal rearrangements with a pathological clone.
Introduction
The tremendous diversity of antigen receptors stems from genetic recombination occurring during early stages of lymphopoiesis. Random assembly of the many variable (V), diversity (D), and joining (J) genes, and pairing of both chains of these heterodimeric receptors provide substantial combinatorial diversity, which is considerably enhanced by the so-called junctional diversity.1,2 Hence, rearrangements of IG or TR genes constitute unique, cell-specific, molecular markers for B and T lymphocytes, respectively. As, in most instances, tumor cells are the progeny of a single transformed malignant cell, analysis of antigen receptor gene rearrangements by PCR and capillary electrophoresis (GeneScan) sizing provides a method for clonality assessment of lymphoid proliferations.3–6
Analysis of T-cell lineage clonality is particularly prone to interpretation issues as clonal rearrangements can be detected in non-malignant conditions including those associated with perturbed and restricted immune repertoires, such as chronic infection,7,8 auto-immune disease,9–11 bone marrow transplantation12 as well as in elderly individuals.13,14 Finally, amplification of IG/TR gene rearrangements from rare B or T-cells in a sample containing few lymphocytes can generate a seemingly clonal profile, termed pseudoclonality.15
Standardization of the molecular detection of lymphoid clonality was achieved almost 15 years ago within a European consortium involving over 45 laboratories (BIOMED-2 Concerted Action BMH4 CT98–3936, hereafter named EuroClonality).16 This resulted in a series of robust and highly reliable, polymerase chain reaction (PCR)-based assays, along with interpretation guidelines, which are now widely used in diagnostic laboratories.15
TRG genes have been a preferential target for T-lineage clonality as (i) they are rearranged in all but the most immature T lymphocytes of both the TR γδ and αβ lineages, and (ii) the limited number of TRGV and TRGJ genes allows their amplification with a small set of primers. The EuroClonality/BIOMED-2 TRG assay was designed as 2 multiplex PCR tubes, each with 2 fluorochromes, hereafter termed TRG-2T-2F. No TRGJP primer was included in order to avoid amplification of invariant, “canonical” TRGV9-TRGJP rearrangements, thus preventing their false identification as a clonal product.17 TRGV primers were positioned in such a way that they allowed TRGV gene identification based on the size of the PCR products (Fig. 1). Labeling the 2 reverse primers with different fluorochromes also permitted distinction of TRGJ1/2 and TRGJP1/2 genes using GeneScan analysis. Therefore, in addition to clonality assessment, this assay could be used for partial TRG genotyping of malignant T-cell populations, a useful feature for subsequent minimal residual disease (MRD) analysis, in particular for acute lymphoblastic leukemia samples.18
Figure 1.
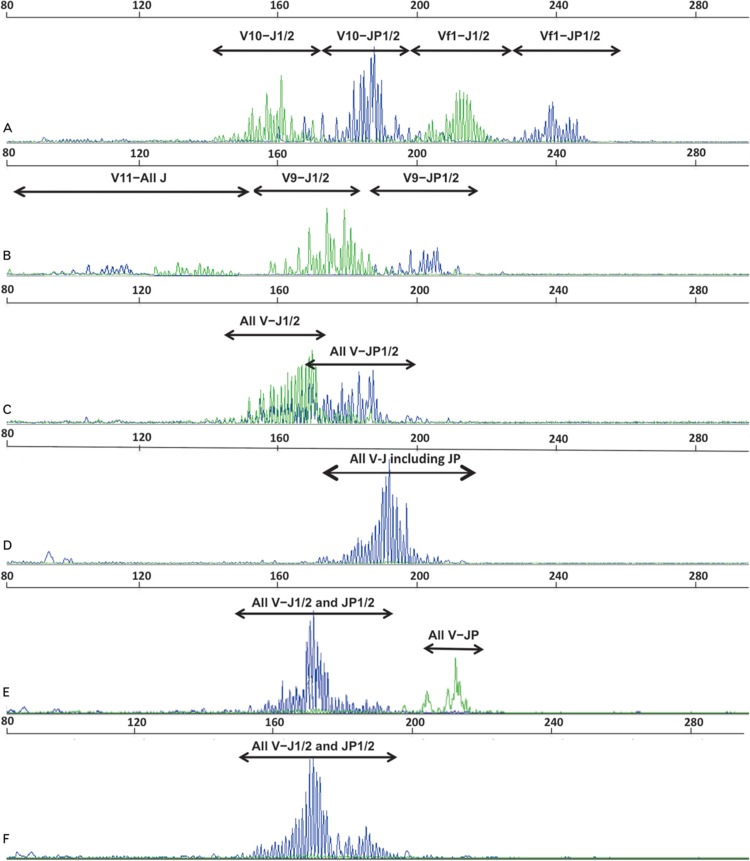
GeneScan analysis of the same polyclonal sample with all PCR assays. (A) TRG-2T-2F tube A; (B) TRG-2T-2F tube B; (C) TRG-1T-2F; (D) IVS-1T-1F; (E) TRG-1T-JPgr with identical FAM (blue) labeling of the TRGJP1/2 and TRGJ1/2 primers and addition of an HEX-labeled TRGJPgreen (JPgr) primer; (F) TRG-1T-1F with identical FAM labeling of theTRGJP1/2 and TRGJ1/2 primers, optimised PCR conditions, and no TRGJP primer.
A drawback of this approach in the context of diagnostic hematopathology is that the PCR products are scattered over a wide size range and cluster according to distinct TRGV-TRGJ combinations. As a consequence, polyclonal T lymphocytes demonstrating rare TRGV-TRGJ rearrangements, for example those using TRGV11, are at risk of being mistaken for a clonal population, due to the absence of a polyclonal background for that type of rearrangement (Fig. 1B).
With these considerations in mind, the EuroClonality consortium undertook to develop an alternative TRG multiplex PCR assay with the following specifications: (i) to regroup PCR products within a limited size range by modifying primer positions in order to avoid over-interpretation of minor peaks of unknown significance, (ii) to combine all primers within a single tube, (iii) to generate relatively short PCR products (<200 bp) to facilitate analysis of FFPE samples in diagnostic pathology laboratories. Several one-tube TRG assays have been described 19–23 and one is commercially available from the Invivoscribe (hereafter termed IVS) company. In the present study, we evaluated this new one-tube EuroClonality TRG-1T-2F assay as well as the one-tube assay from IVS (IVS-1T-1F) on a large series of T-cell malignancies and reactive samples, in comparison with the conventional TRG-2T-2F assay.
Results
Primer positions and amplicon sizes of the different TRG PCR assays
Typical GeneScan profiles obtained on a polyclonal sample are shown for all tested TRG multiplex PCR assays in Figure 1. PCR products obtained with TRG-2T-2F primers were dispersed over 8 size ranges, that is, 145 to 255 bp for tube A and 80 to 220 bp for tube B, as described.16 The TRG-1T-2F assay resulted in 2 distinct overlapping Gaussian curves depending on TRGJ usage, with a 140 to 200 bp range. A single Gaussian curve, ranging from 160 bp to 210 bp was observed with the IVS-1T-1F system.
Sensitivity of the various TRG PCR assays
Analytical sensitivity was determined by testing DNA dilutions (10%, 5%, and 1%) from 7 human T-cell lines in peripheral blood mononuclear cells from a healthy donor. Depending on the position of the clonal rearrangement(s) within the Gaussian curve of polyclonal peaks, the sensitivity threshold varied from 1% to 10% (SDC Table 1, Supplemental Digital Content). Two cell lines (HSB2 and Jurkat) had a slightly increased sensitivity with the TRG-2T-2F PCR since their clonal rearrangements were situated outside the bulk of the Gaussian distribution of polyclonal rearrangements, particularly evident with the more dispersed repertoires in this assay.
Inter-laboratory comparison of the GeneScan profiles of the different TRG PCR assays
GeneScan profiles for all 3 PCR assays were first compared between the paired laboratories and led to concordant interpretations in both laboratories in 205/239 (86%) cases. In further 7 cases, the GeneScan profiles were actually similar, but minor peaks of unclear significance with the TRG-2T-2F PCR (data not shown) were interpreted differently in the 2 laboratories. Joint re-analysis of the GeneScan profiles enabled the 2 laboratories to systematically come to a consensus, essentially by not over-interpreting minor peaks.
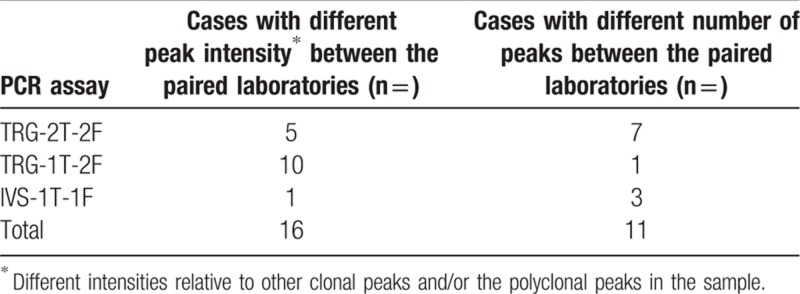
In the remaining 27/239 cases (11%), the GeneScan profiles differed between paired laboratories, either regarding the intensity of peaks (n = 16) or the number of peaks (n = 11) (Table 1). Differences in peak intensities were mostly due to the instrument settings. The paired laboratories came to a consensus conclusion upon re-analysis of their data, after taking into account this instrument-related bias, with the balance between the intensity of the 2 fluorochromes differing in a systematic fashion between laboratories (Fig. 2A). When 2 laboratories found a different number of peaks for a given PCR assay (Fig. 2B), it was analyzed by a third laboratory (Paris-Pitié) and the profile found in 2 out of 3 laboratories considered to be the consensus one (see below).
Table 1.
Inter-laboratory discrepancies of the different TRG PCR assays
Figure 2.
Inter-laboratory discrepancies regarding peak intensities in the TRG-1T-2F assay. (A) Difference of the green dye peak intensity (arrow) compared to the blue one. (B) Discrepancy regarding the detection of the blue dye peak (arrow). ALCL, anaplastic large-cell lymphoma.
Of note, neither type of discrepancies (number or intensity of peaks) caused a change of conclusion for the clonality status, since they only concerned the number of rearranged alleles (clonal population with either mono-allelic or bi-allelic rearrangements) or the presence of a minor peak of unknown significance in addition to a major clonal peak. Inter-laboratory discrepancies were solved for all these 27 samples after this first step of data reviewing, although there was clear inter-laboratory heterogeneity in the relative signal intensity of fluorochromes.
Inter-assay comparison of the GeneScan profiles for the 3 PCR assays
The 3 PCR assays were then compared based on the consensus profile for each sample. Results were concordant between the 3 PCR assays in 155/239 cases (65%). Among the 84 remaining samples, the one “outlier” PCR assay was considered to be discordant with the other 2.
In 61/84 samples this discordance had no impact on the overall interpretation. As detailed in SDC Figure 1 (Supplemental Digital Content), such discrepancies included bi-allelic vs mono-allelic rearrangement (45/61 cases) or cases with additional minor peaks (16/61), all of which were seen with the TRG-2T-2F assay. The outlier assay was IVS-1T-1F in 26 cases, TRG-2T-2F in 20 cases, and TRG-1T-2F in 15 cases. Overall, the 3 PCR assays provided similar conclusions for 216 samples (90%).
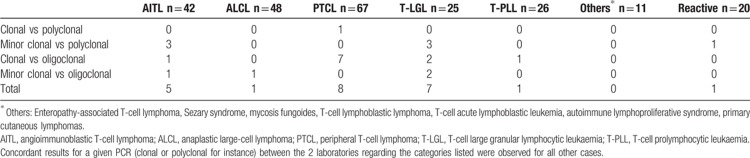
For the 23 other samples (10% of the cohort), the discordant results between PCR led to a change of conclusion depending on which assay was considered (SDC Fig. 1, Supplemental Digital Content). These were classified into 4 categories: clonal vs polyclonal (n = 1), minor clonal vs polyclonal (n = 7), clonal vs oligoclonal (n = 11) and minor clonal vs minor oligoclonal (n = 4) (Table 2 and SDC Table 2, Supplemental Digital Content). All but one of the reactive samples were concordant between the 3 PCR assays. Only 1 PTCL sample (DE-002) displayed a significant change of conclusion, switching from polyclonal to clonal. Of note, this case had previously been shown to have a clonal TRB gene rearrangement pattern.24 Taken together, the outlier discordant assay was IVS-1T-1F in 10 cases, TRG-1T-2F in 9 cases and TRG-2T-2F in 4 cases (SDC Fig. 1 and SDC Table 2, Supplemental Digital Content).
Table 2.
Discrepancies between the interpretation of TRG-2T-2F, TRG-1T-2F, and IVS-1T-1F PCR assays and their consequences with respect to the conclusion according to disease category
Technical improvements of the EuroClonality TRG-1T-2F PCR assay
Given that a frequent source of discordant results occurring both in inter-laboratory TRG-1T-2F duplicates (Fig. 2) and between the 2 one-tube assays (SDC Fig. 1, Supplemental Digital Content) concerned the number of dominant clonal peaks (1 vs 2), we investigated the basis of these discrepancies. This resulted from consistent differences in the balance between the 2-color fluorescent PCR products (Fig. 2). We, therefore, changed to single fluorescent (FAM) labeling of both TRGJG1/2 and TRGJP1/2 primers. Another possible explanation for the difference in peak number between the IVS-1T-1F and TRG-1T-2F assays was the lack of a TRGJP primer in the latter. To test this hypothesis, a modified TRG-1T-JPgr assay, which included a differently labeled (HEX/green) TRGJP primer, was tested together with the initial TRG-1T-2F and the IVS-1T-1F assays by 2 laboratories (Paris-Pitié, Paris-Necker) on an additional series of 19 DNA samples (SDC Table 3, Supplemental Digital Content). These cases were selected from local archives for the presence of canonical (n = 12) or pathological (n = 7) TRGJP rearrangements previously identified with in-house multiplex TRG PCR assays including a TRGJP primer. As expected, all cases displayed pathological or canonical TRGJP rearrangements that were not detected by the initial TRG-1T-2F PCR (data not shown), demonstrating the lack of cross-amplification of TRGJP rearrangements by the TRGJ1/2 or TRGJP1/2 primers. For the 7 cases with a pathological TRGJP rearrangement, the new TRG-1T-JPgr assay did not change the clonality status since all of them also had a second clonal non-TRGJP rearrangement, which was clearly detected by the initial TRG-1T-2F assay. The IVS-1T-1F PCR also amplified both alleles in these cases.
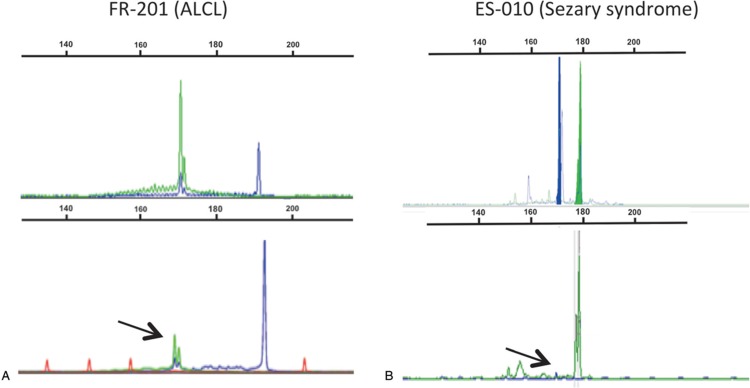
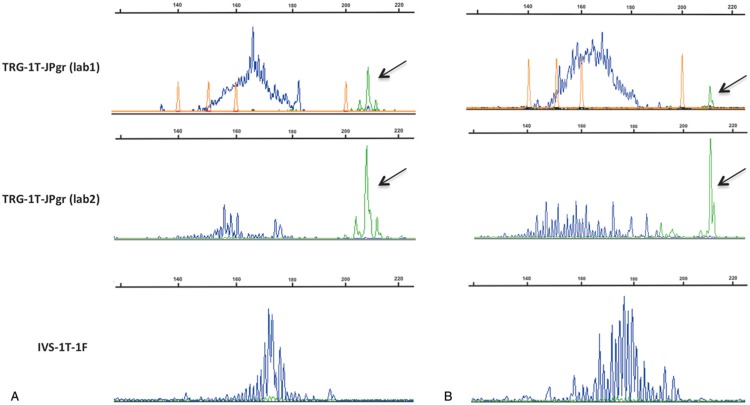
One potential risk of including the TRGJP primer within a single color, single Gaussian distribution as developed by IVS could be the erroneous identification of a canonical TRGV9-TRGJP rearrangement as evidence of a pathological clonal population. However, this did not prove to be the case, since the 12 samples with non-malignant TRγδ populations did not generate clonal peaks upon testing with the IVS-1T-1F PCR assay. In contrast, we found that the dual-labeled TRG-1T-JPgr profiles with the canonical rearrangements outside the Gaussian curve could sometimes be difficult to interpret (SDC Fig 2, Supplemental Digital Content) and inter-laboratory comparison showed striking differences in the relative intensity of the HEX- and FAM-labeled PCR products, whereby one laboratory under-estimated and the other over-estimated the TRGJPgr repertoire (Fig. 3).
Figure 3.
Profiles of the TRG-1T-JPgr PCR assay in two cases (A and B) from a separately selected cohort of samples with canonical TRG-JP rearrangements. Difference in fluorochrome intensity between two laboratories (top and middle panels) identifies a potentially misleading over-amplification of the green dye in the second laboratory (middle panel). In the bottom panel, profiles of the IVS-1T-1F PCR assay are shown for reference. Canonical JP rearrangements (in green) are indicated by arrows.
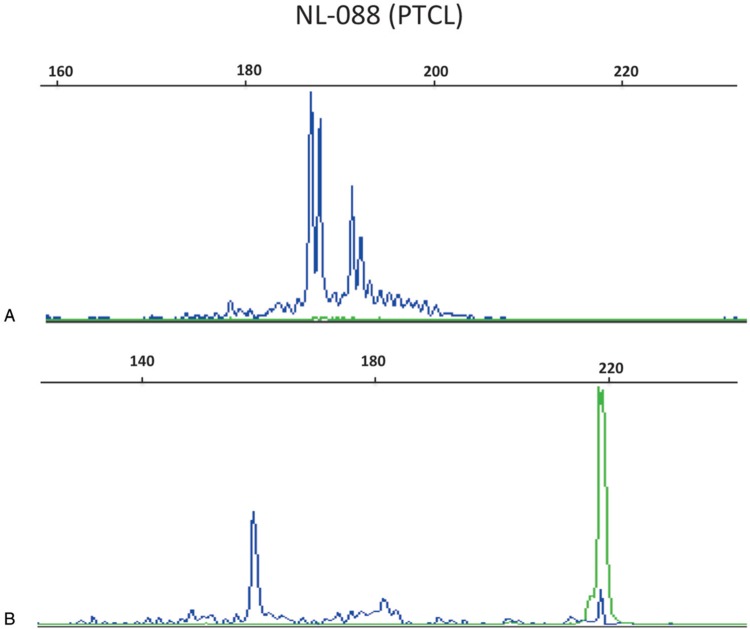
This TRG-1T-JPgr assay was then further tested on 17 of the 26 cases associated with an extra peak with IVS-1T-1F compared to TRG-1T-2F (but with no discrepancy for overall conclusion) (SDC Fig 1, Supplemental Digital Content). Only one sample (NL-088, a PTCL) showed a TRGJP rearrangement (Fig. 4), indicating that the vast majority of discrepancies were not due to the absence of the TRGJP primer in the initial TRG-1T-2F assay. Of note, NL-088 also demonstrated a second weaker non-TRGJP clonal peak. Taken together, the risk of potential false-positive results generated by inclusion of a TRGJPgr primer generating larger PCR products was considered to outweigh the negligible risk of false-negative results when using a TRG multiplex which does not allow detection of TRGJP rearrangements.
Figure 4.
Genescan profiles of case NL-088 (PTCL) displaying a clonal TRGJP rearrangement. (A) IVS-1T-1F PCR assay with 2 clonal peaks. It is not possible to identify which peak corresponds to TRGJP rearrangement with this PCR; (B) TRG-1T-JPgr PCR assay with a TRGV-TRGJ rearrangement in blue and a pathological TRGJP rearrangement in green.
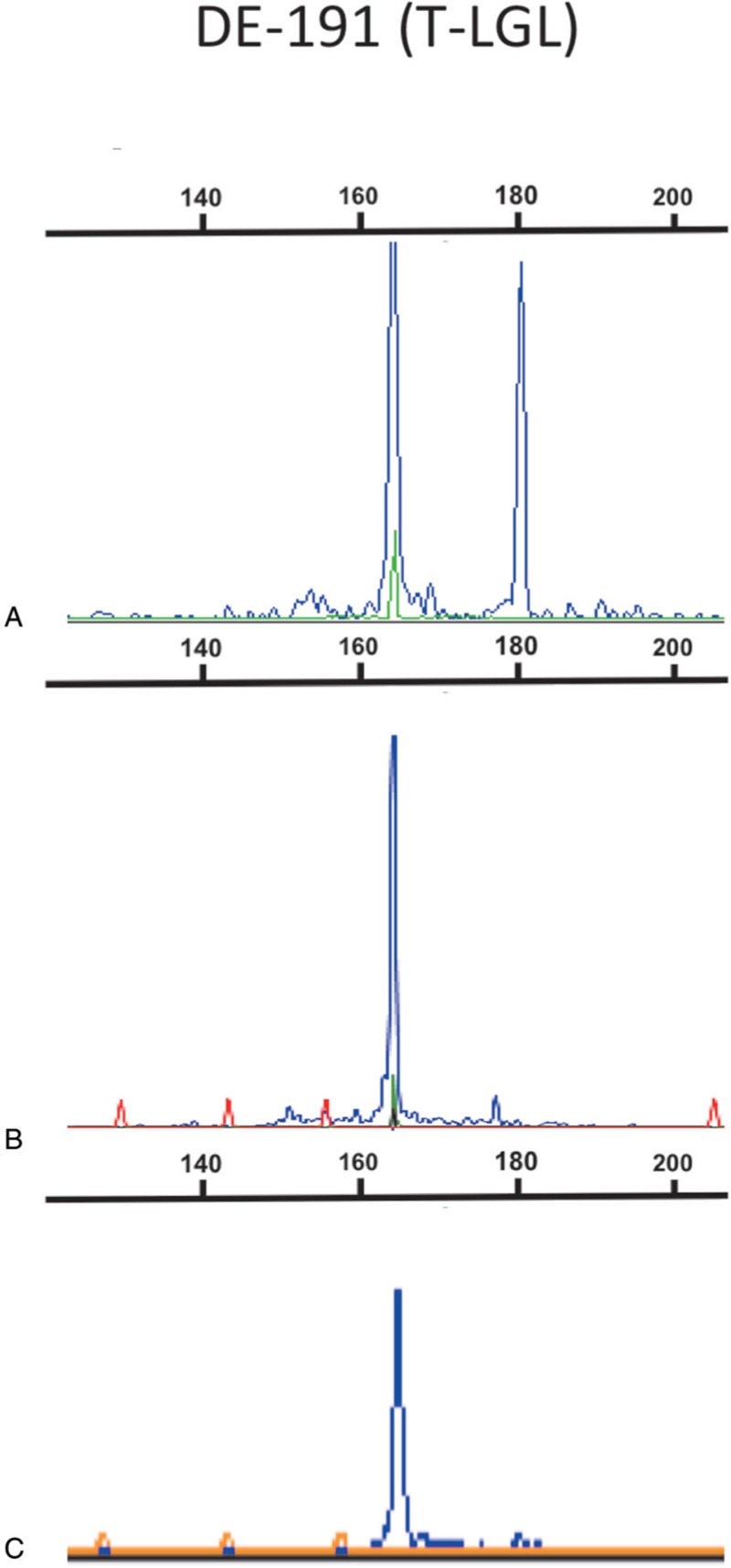
In addition to this primer evaluation, a variety of parameters (annealing temperature, dNTPs, magnesium concentration, type of thermocycler, etc.) were investigated. This identified the dNTPs source as a major determinant causing discrepant results in the size and ultimately the number of clonal peaks detected (Fig. 5). Discordance occurred in 12/28 (43%) samples tested (see below) but was solved when using dNTPs from the same commercial source. A final, optimized TRG-1T-1F PCR assay taking all these parameters into account and devoid of a TRGJP primer was then adopted, as described in the Materials and Methods section. It differed from the TRG-1T-2F essentially by use of a single fluorochrome for TRGJ primers and optimized dNTPs.
Figure 5.
Impact of dNTP reagents on GeneScan profiles. TRG-1T-2F PCR assay profiles of sample DE-191 (T-LGL), analyzed in 3 laboratories (A, B and C) using the same PCR conditions, except for the dNTP source. Laboratories B and C used the same dNTP reagents resulting in a single peak, while laboratory A used dNTP reagents from a different provider (ThermoFischer), with the PCR profile displaying 2 clonal peaks.
Evaluation of the optimized EuroClonality TRG-1T-1F PCR assay
The optimized TRG-1T-1F assay was evaluated on 28 samples with sufficient available material from the original cohort of 60 samples which had demonstrated clear discrepancies between the number of peaks detected with the initial TRG-1T-2F and IVS-1T-1F assays (SDC Fig 1, Supplemental Digital Content). This comparison was performed in 3 laboratories (Paris-Pitié, Paris-Necker, Erasmus MC) and the combined results are shown in Table 3.
Table 3.
Evaluation of the EuroClonality TRG-1T-1F assay on 28 of the 60 samples initially showing a discrepancy in the number of alleles between IVS-1T-1F and TRG-1T-2F assay
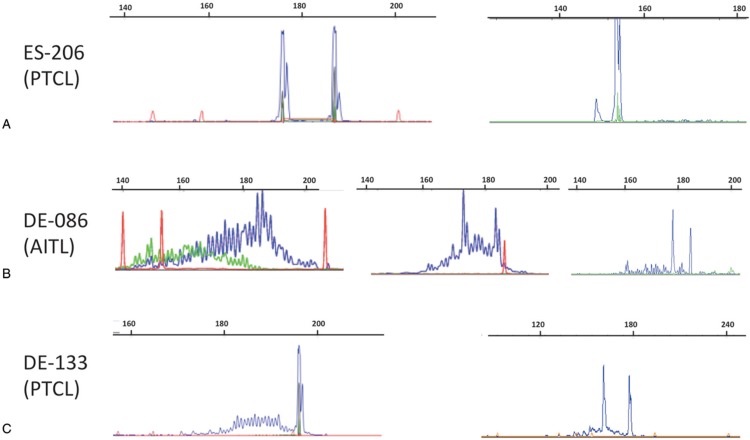
Of the 17 samples that initially had an extra peak in the IVS-1T-1F assay, all but 2 (DE-167 and NL-088) now displayed the same number of alleles between TRG-1T-1F and IVS-1T-1F. They included 3 cases (ES-127, ES-206, NL-096) with 2 virtually overlapping peaks in the TRG-1T-1F GeneScan profile (Fig. 6A) and 2 (DE-084 and DE-086) which were initially classified as polyclonal with the TRG-1T-2F while IVS-1T-1F showed 1 or 2 minor peaks above the polyclonal background. These minor peaks became apparent with the definitive TRG-1T-1F PCR (Fig. 6B). Since the NL-088 discrepancy was due to the absence of a TRGJP primer in the TRG-1T-2F PCR mix (Fig. 4), only one case (DE-167) remained truly discordant.
Figure 6.
Examples of discrepancies between one-tube TRG PCR assays. (A) Case ES-206 (PTCL) with bi-allelic rearrangements with the IVS-1T-1F PCR (left) and apparently only 1 rearrangement but possibly with 2 overlapping peaks in the TRG-1T-1F PCR (right). (B) Case DE-086 (AITL) with a polyclonal profile with the TRG-1T-2F PCR (left), and 2 peaks on a polyclonal background with both the IVS-1T-1F (middle) and TRG-1T-1F assays (right). (C) Case DE-133 (PTCL) with bi-allelic rearrangements with TRG-1T-1F PCR (right) and only 1 rearrangement but possibly with 2 overlapping peaks in the IVS-1T-1F PCR (left). Red peaks correspond to size markers.
Conversely 11 samples initially had an extra peak with TRG-1T-2F compared to IVS-1T-1F. In contrast to the previous category, the use of the TRG-1T-1F PCR mix did not change these discrepancies. However, upon careful examination of the IVS-1T-1F GeneScan profiles, 6 cases had 2 overlapping peaks (Fig. 6C). As such, only 5 of the 11 cases had true discordant profiles (Table 3).
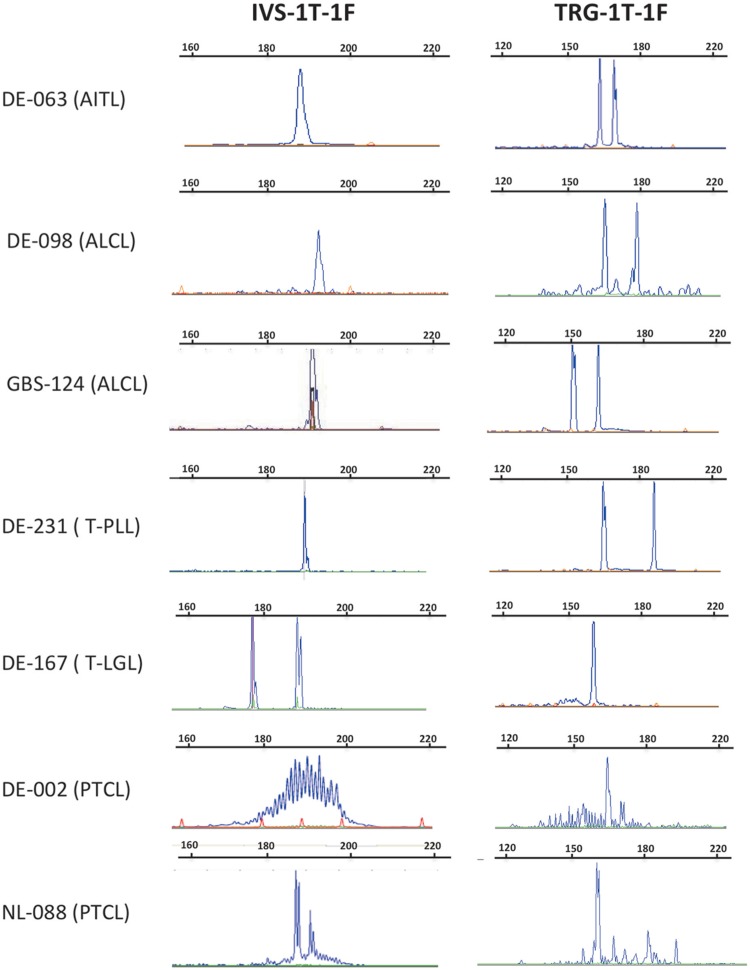
Overall, 7/28 tested samples remained discordant between the 2 one-tube assays with (i) a second allele not being detected by the TRG-1T-1F assay in 2 samples (DE-167 and NL-088) and by the IVS-1T-1F assay in 4 samples (DE-063, DE-098, DE-231, GBS-124), and (ii) a clonal, mono-allelic rearrangement in a PTCL sample (DE-002) detected by the TRG-1T-1F but not the IVS-1T-1F assay (Table 3 and Fig. 7). As mentioned above, this case was demonstrated to be clonal by analysis of the TRB locus 24. For sample NL-088, with 2 clonal rearrangements with the IVS-1T-1F PCR, the lack of detection of the second clonal peak by the TRG-1T-1F PCR was due to the absence of a TRGJP primer in this assay (Fig. 4). Therefore only 1 of these 7 samples (DE-002) was associated with a conclusion discrepancy (clonal vs polyclonal).
Figure 7.
GeneScan profiles of cases with discrepancies between IVS-1T-1F (left) and TRG-1T-1F (right) assays. Regarding case NL-088, the TRGV-TRGJ rearrangement was seen in both PCRs but the TRGV-TRGJP rearrangement was not detected with the TRG-1T-1F PCR, due to the absence of a TRGJP primer, in contrast to the assay in Figure 4. It is not possible to determine which peak corresponds to the TRGJP rearrangement in the IVS-1T-1F PCR, as the primer information is not provided in the IVS TRG V2.0 kit assay.
Since only 7 (25%) of the 28 discordant DNAs analyzed with the optimized TRG-1T-1F PCR remained discordant, if we extrapolate this to the total 60 discordant samples, we can estimate an overall 6% discordancy rate (15/239) from the initial EuroClonality cohort between the optimized TRG-1T-1F and IVS-1T-1F PCR.
In conclusion, the excessive dispersion of the two-tube TRG-2T-2F PCR generated minor peaks, with a risk of false-positive results. TRG-1T-1F and IVS-1T-1F assays gave comparable results, with a slightly more dispersed multiplex in the TRG-1T-1F assay, leading to a lower risk of superposition of biallelic clonal rearrangements, and an intentional failure to detect both canonical and clonal TRGJP rearrangements.
Discussion
The assessment of T-cell clonality is often based on the molecular analysis of TR gene rearrangement patterns, as in contrast to B cell proliferations, one cannot rely upon the immunophenotypic detection of “monotypic” TR chain restriction. Interpretation of TR clonality is, however, often complicated by detection of minor clones which can reflect perturbations of the immune repertoire rather than neoplastic T-cell populations.7–9,11,13,14
The initial TRG-2T-2F PCR assay was designed to allow both clonality detection and partial VJ typing for target identification in MRD strategies, with significant dispersion of 8 sub-repertoires according to TRGV and TRGJ usage. This inevitably led to the higher risk of false-positive results, particularly for the minor sub-repertoires and in samples with few T lymphocytes. We therefore developed a preliminary single-tube TRG-1T-2F PCR optimized for clonality assessment, including in FFPE samples. This assay was compared to the original TRG-2T-2F PCR and the commercial IVS-1T-1F kit, in a multi-center study including 10 EuroClonality laboratories, on 239 samples from the previous BIOMED-2 report.24 Results showed that the vast majority (86%) of samples were concordant, but 14% displayed discrepant profiles, including after panel review.
Additional peaks were often observed with the TRG-2T-2F system due the dispersion of PCR size products, particularly for tube B, where seemingly clonal peaks could appear due to the low number of T-cells using more rare TRGV-TRGJ combinations. Discrepancies between PCR assays were also due frequently to the variable presence of a second clonal peak resulting from clear inter-laboratory differences in detection of the relative intensity of the 2 fluorochromes, presumed to be due to differences in instrument choice and settings. In order to avoid this problem, it was considered preferable to use a single fluorochrome, as is the case with the IVS-1T-1F PCR and our optimized TRG-1T-1F assay. These conclusions are in keeping with previous studies, 20,25,26 in favor of homogeneously labeled PCR products with limited size range. Cushman-Vokoun et al. reported that their one-tube assay was as sensitive but more specific than the BIOMED-2 TRG-2T-2F, and underlined the difficulty of interpreting clonal peaks when rearrangements are distributed over several separate areas.20 By splitting exploration of the TRG locus over 4 tubes, Patel et al. confirmed the risk of overinterpretation of “pseudoclonality” and favored the combination of several assays, including TRB assessment.25 The concern with multi-distribution design assays was also underlined by Kansal et al. comparing the TRG-2T-2F assay with a next-generation sequencing-based one-tube assay. 26
Further evaluation showed that most discrepancies were due to failure to amplify the second clonal peak with PCRs using inappropriate dNTPs, demonstrating that minor technological modifications can significantly impact on clonality profiles. The role of dNTPs as a critical parameter in multiplex PCR has been previously reported,27,28 with these components appearing to be particularly sensitive to repeated thawing/freezing cycles. With the optimized TRG-1T-1F assay, the concordance rate between the 2 one-tube TRG PCR assays reached 94%. The capacity of the EuroClonality group to perform widespread, multicenter testing allows identification of minor details in technical parameters which can lead to discordant inter-laboratory results, as demonstrated here for the balance between fluorochromes and dNTP usage.
The simplicity of the TRG combinatorial repertoire allows inclusion of primers for all rearrangements with a limited risk of false-negative results. However, there is a risk of a false-positive result in the presence of canonical TRGV9-JP rearrangements, which generate PCR products of uniform size but which cannot be easily detected in single color multiplex strategies with overlapping repertoires. The TRGJP gene-specific primer was, therefore, intentionally omitted from the TRG-2T-2F and TRG-1T-1F primer sets. This led to the expected failure to detect a clonal TRGJP rearrangement in one of the 239 samples, in contrast to the IVS-1T-1F assay, which does include a TRGJP primer that generates rearranged amplicons overlapping with the other sub-repertoires. We, therefore, tested a modified TRG-1T-JPgr PCR which included a TRGJP primer generating slightly larger PCR products. This proved, however, to be potentially problematic since it was not easy to distinguish canonical from pathological TRGJP rearrangements. In order to avoid misinterpretation of these rearrangements in not very experienced laboratories, we therefore chose to remove the TRGJP primer from the final, optimized, TRG-1T-1F assay. Cushman-Vokoun et al. included a TRGJP primer in their assay, which places TRGV-JP rearrangements in the middle of the Gaussian curve, thereby decreasing -but not eradicating- the risk of false positivity.20 They tested it on a relatively limited cohort of 40 samples, and found a good specificity, with no false positives. Presuming that this PCR assay is similar or identical to the IVS-1T-1F PCR used here, we confirmed the absence of false positives in all 12 DNA samples with canonical TRGJP rearrangements. We consider, however, that this at least theoretical risk outweighs the advantage of including a TRGJP primer, except in experienced laboratories. If a TRGJP primer is to be used, it should be one which has been shown to give satisfactory results when placed within the Gaussian distribution, as in Cushman-Vokoun et al., or which has been placed outside the Gaussian curve and differentially labelled, as in Derrieux et al.29 Indeed, in experienced laboratories, the identification of canonical VG9-JP rearrangements can provide a useful “positive control”. They are essentially detected in peripheral blood, so the potential risk of false positive results in tissue diagnostics is limited. In our series, which included both blood and tissue samples, only 1/239 (0.4%) was found to have a clonal TRGJP rearrangement, and this case displayed also a second, albeit minor, clonal peak. Considering that biallelic rearrangements are reported in more than half of T-cell malignancies, and TRGJP rearrangements in about 3% of cases,21 exclusion of this primer will lead to a very low risk of false-negative results. Furthermore, in cases with unexpected negativity when a clonal T-cell proliferation is strongly suspected, TRG clonality analysis should be complemented by TRB clonality evaluation, as recommended by the EuroClonality/Biomed2 guidelines.15,16,24
One-tube TRG PCR assays present several advantages relative to the two-tube TRG-2T-2F assay, including simplicity, reduced amount of DNA, ease of interpretation, lower risk of false-positive results and improved applicability to DNA extracted from FFPE fixed samples due the small size of the amplicons. Unfortunately, many (19/28) of the FFPE samples from the initial Biomed2 cohort were degraded, limiting the value of the analysis of the remaining 9 FFPE samples. Derrieux et al. have, however, recently demonstrated the superiority of a TRG-1T-JPgr variant (with NED labeling of the TRGJP primer) compared to an in-house 2-tube TRG.29 All these advantages are shared between the IVS-1T-1F and the TRG-1T-1F assays, as they gave largely comparable results. The 2 main differences between the profiles generated by these PCR systems are (i) the slightly more dispersed Gaussian distribution of the TRG-1T-1F PCR (see Fig. 1) and (ii) the aforementioned inclusion of a TRGJP primer in the IVS-1T-1F assay. As could be anticipated from the smaller Gaussian size distribution of the IVS-1T-1F PCR, a small number of clonal samples (15/239 in this series) appeared to have a mono-allelic rearrangement using this assay but bi-allelic rearrangement with the TRG-1T-1F, usually due to superposition of both clonal products in the IVS-1T-1F PCR. This is not, however, a practical problem in standard hematopathology practice.
Several studies have reported that high-throughput sequencing methods can be used for T-cell clonality determination.26,30–32 These techniques are currently more labor-intensive, longer and expensive. They also require dedicated bioinformatics pipelines and lack standardization for clinical applications. Conventional assays based on PCR and capillary electrophoresis therefore still have a place of choice in the molecular diagnostic tools for lymphoid clonality assessment, at least for the near future.
In conclusion, we describe a new simple, robust multi-center validated single-tube TRG PCR assay, which will lead to greater inter-laboratory reproducibility. It shows similar performance to the commercial one-tube IVS TRG assay when compared to the original two-tube TRG-2T-2F PCR, thus validating the use of both the EuroClonality TRG-1T-1F and IVS-1T-1F assays in T-cell clonality assessment, when tested over a wide range of T-cell malignancies. It remains possible that one or other 1T-1F (IVS or EuroClonality TRG) PCR may prove to be preferable in particular T lymphoid malignancies and they may be complementary.
Materials and methods
Primer design of the initial one-tube EuroClonality TRG assay
The EuroClonality/BIOMED2 one-tube preliminary system (TRG-1T-2F) consisted of a single PCR reaction with all 4 TRGV primers (TRGV1f, TRGV9, TRGV10, and TRGV11) and 2 TRGJ primers: TRGJ1/2 and TRGJP1/2. As for the original two-tube EuroClonality/BIOMED-2 (TRG-2T-2F) assay, these reverse TRG-1T-2F primers were initially labeled with different fluorochromes, with TRGJ1/2 HEX-labeled (green) and TRGJP1/2 FAM-labeled (blue), but in the final assay (TRG-1T-1F) only one-color labeling (FAM) was retained for both primers (see below). The sequences of the primers and their positions on the TRGV and TRGJ genes compared to TRG-2T-2F assay are depicted in SDC Table 4 (Supplemental Digital Content).
The primers were first tested and validated in Paris-Necker Hospital, then centrally produced and aliquoted in Erasmus MC, Rotterdam to ensure that all participating laboratories used identical primers. Of note, the sequences of the primers used in the IVS-1T-1F PCR assay (T-Cell Receptor Gamma Gene Rearrangement Assay 2.0 kit, www.Invivoscribe.com) are not available. It is, however, noteworthy that the TRG-1T-2F, TRG-1T-1F, and TRG-2T-2F assays do not include a TRGJP primer, whereas the IVS kit does.
Sample collection
The 261 samples (233 fresh/frozen and 28 FFPE) used in this study had been previously collected and thoroughly evaluated for T-cell clonality within the work packages of EuroClonality/BIOMED-2 Concerted Action BMH4 CT98–3936.16,24,33 DNA aliquots were taken from archival material collected and analyzed in these BIOMED-2 studies. From this initial collection, 22 samples were later excluded because of poor DNA quality as evidenced by the absence of amplification with all PCR assays. A total of 239 samples (including 230 fresh/frozen and 9 FFPE) with adequate DNA quality were finally retained for a two-by-two comparative analysis representing true duplicates by the 10 participating laboratories (AP-HP, Paris-Necker; AP-HP, Paris-Pitié; AP-HP, Paris-Saint-Louis; AZ Delta, Roeselare; HMDS, Leeds; Charite, Berlin; UniKiel, Kiel; IPO-Lisboa, Lisbon; Radboud UMC, Nijmegen; Erasmus MC, Rotterdam). The samples included 42 angioimmunoblastic T-cell lymphoma (AITL), 48 anaplastic large-cell lymphoma (ALCL), 26 T-cell prolymphocytic leukemia (T-PLL), 25 T-cell large granular lymphocytic leukemia (T-LGL), 67 peripheral T-cell lymphoma (PTCL), 11 other T-cell malignancies (1 Enteropathy-associated T-cell lymphoma, 1 Sezary syndrome, 1 mycosis fungoides, 1 T-cell lymphoblastic lymphoma and 3 T-cell acute lymphoblastic leukemia (T-ALL), 2 autoimmune lymphoproliferative syndrome (ALPS), 2 primary cutaneous lymphomas) as well as 20 reactive samples. The vast majority of AITL, ALCL, PTCL, AILD and reactive samples were from fresh-frozen tissue and the T-PLL, T-LGL and T-ALL/LBL samples were from blood or bone marrow. Four fresh-frozen skin and one gut biopsies were also analyzed. Histopathological categories were those established by a panel review of experienced hematopathologists at the time of the BIOMED-2 studies, as specified in the initial publications.16,24,33 An additional 19 clinical samples submitted for clonality analysis from patients from Paris-Pitié and Paris-Necker hospitals were added specifically since they had a known TRGJP rearrangement, as detected by in-house multiplex PCR assays. All samples had been obtained with informed consent from the patients according to each participating institution.
Cell lines
Seven cell lines (HSB-2, Jurkat, RPMI 8402, CCRF-CEM, HPB-ALL, MOLT-3, and SU-DHL-1) obtained from commercial sources (https://www.atcc.org; https://www.dsmz.de) and known to be positive for TRG rearrangements were used as positive controls. To assess the sensitivity of the PCR assays, the DNA of each cell line was diluted at 10%, 5%, and 1% in peripheral blood lymphocytes DNA from a healthy donor.
PCR studies
All PCR reactions were carried out using 50 ng of DNA. For amplifications with TRG-1T-1F and TRG-2T-2F primers, the PCR reactions were largely performed according to the EuroClonality/BIOMED-2 protocol.16 Based on the present work, we propose an optimized TRG-1T-1F PCR assay. The reaction conditions were set for a final volume of 25 μl with 2.5 μl of ABI Gold Buffer 10X, 1U of AmpliTaq Gold (ThermoFischer Scientific, Waltham, MA) and 5 pmol of each primer. MgCl2 and dNTP (ThermoFischer Scientific) were used at a final concentration of 2.5 mM and 200 μM respectively. Cycling conditions started with a first step at 94°C for 10 minutes followed by 35 cycles including denaturation (1 minute at 94°C), annealing (1 minute at 60°C), extension (1.5 minute at 72°C) and a final extension at 72°C for 10 minutes. PCR reactions using the IVS-1T-1F assay were performed according to the manufacturer's recommendations. For all systems, PCR products were analyzed by GeneScan profiling according to the EuroClonality recommendations (www.euroclonality.org).16 Briefly, cases were called clonal when displaying 1 or 2 dominant peaks with no or weak polyclonal background, and oligoclonal when there were 3 or more peaks. They were considered as displaying minor clonal or minor oligoclonal profiles when clonal peaks (respectively 1–2, or 3 or more) were observed on a polyclonal background, but with a clonal peak height a least twice that of the polyclonal background.34 Results were reported according to EuroClonality guidelines.15
Organization of the workflow and review of data
A total of 10 molecular diagnostic laboratories from 6 European countries participated in the validation study. Each DNA sample was tested with the 3 PCR systems by 2 different laboratories. Each paired laboratory received the same 50 to 55 DNA samples to mimic duplicate analysis. Data were first compared between paired laboratories. Some discrepant results, due to sample inversion, different instrument settings or divergent interpretation were solved at this stage. All of the revised data including GeneScan profiles and scoring sheets were then collected in one center (Paris-Pitié) and reanalyzed. When agreement could not be obtained between the paired laboratories regarding the molecular profile for a given PCR assay, the sample was sent to Paris-Pitié laboratory for a third evaluation with the discordant PCR assay. Thus, the final interpretation for these cases included similar duplicate results from at least 2 of the 3 laboratories, which was considered to be the consensus profile.
The next step of the data review concerned the inter-assay discrepancies per sample. Discrepant results between PCR systems could lead to change of conclusion. Each time, the outlier PCR system was reported. Inter-assay discrepancies could also be consequence-free, that is, when a clonal population was found to have a mono-allelic rearrangement with one PCR and bi-allelic rearrangements with another, or when a minor clone was found in addition to a strong clonal rearrangement with one PCR while only the major clone was seen with another PCR.
EuroClonality assay optimization and complementary cohort
An adapted one-tube assay (TRG-1T-JPgr) was used to evaluate the consequences of including the TRGJP primer in the PCR assay. It corresponds to a single-tube fluorescent multiplex PCR adapted from the initial TRG-1T-2F PCR by identical FAM labeling of the TRGJP1/2 and TRGJ1/2 primers and addition of an HEX-labeled TRGJP primer (SDC Table 4, Supplemental Digital Content), adapted from a similarly modified EuroClonality assay which varied by 5’NED labeling of the TRGJP primer.29 The 4 TRGV primers were identical to those in the TRG-1T-2F assay. The TRGJP reverse primer was intentionally placed further from the 5’ end of the TRGJP gene, relative to the positions of the TRGJ1/2 and TRGJP1/2 reverse primers in order to generate larger, distinctly labeled PCR products, favoring detection of canonical TRGV9-TRGJP TRγδ repertoires, which appears as a major peak at 207 bp, flanked by minor peaks at 204 bp and 210 bp (Fig. 1E).
A complementary cohort of 19 stored DNA samples previously obtained from patients with known TRGJP rearrangements identified during routine analysis by in-house TRG PCR assays in Paris-Pitié (n = 9) and Paris-Necker (n = 10) was added for the TRGJP study. These samples were assessed by the IVS-1T-1F and the TRG-1T-JPgr assay in both Paris-Pitié and Paris-Necker, as for the initial cohort, using identical reactions and cycling conditions to the initial EuroClonality TRG-1T-2F assay.
The final EuroClonality TRG-1T-1F assay is similar to TRG-1T-JPgr assay but without the TGRJP primer, and is, as such, a one-tube, one-color assay. Troubleshooting procedures are proposed in SDC Table 5 (Supplemental Digital Content).
Supplementary Material
Footnotes
Citation: Armand M, Derrieux C, Beldjord K, Wabeke T, Lenze D, Boone E, Bruggemann M, Evans PAS, Gameiro P, Hummel M, Villarese P, Groenen PJTA, Langerak AW, Macintyre EA, Davi F. A New and Simple TRG Multiplex PCR Assay for Assessment of T-cell Clonality: A Comparative Study from the EuroClonality Consortium. HemaSphere, 2019;00:00–00. http://dx.doi.org/10.1097/HS9.0000000000000255.
KB, DL, EB, MB, PAE, PG, MH, PV, PJG, AWL, EAM, and FD are members of the EuroClonality corsortium which receives royalties from Invivoscribe for kits based on the EuroClonality/BIOMED-2 Concerted Action BMH4-CT98-3936.
The authors have indicated they have no potential conflicts of interest to disclose.
References
- 1.Tonegawa S. Somatic generation of antibody diversity. Nature. 1983;302:575–581. [DOI] [PubMed] [Google Scholar]
- 2.Davis MM, Bjorkman PJ. T-cell antigen receptor genes and T-cell recognition. Nature. 1988;334:395–402. [DOI] [PubMed] [Google Scholar]
- 3.van Dongen JJ. Analysis of immunoglobulin genes and T cell receptor genes as a diagnostic tool for the detection of lymphoid malignancies. Neth J Med. 1987;31:201–209. [PubMed] [Google Scholar]
- 4.Gleissner B, Maurer J, Thiel E. Detection of immunoglobulin heavy chain genes rearrangements in B-cell leukemias, lymphomas, multiple myelomas, monoclonal and polyclonal gammopathies. Leuk Lymphoma. 2000;39:151–155. [DOI] [PubMed] [Google Scholar]
- 5.Sen F, Vega F, Medeiros LJ. Molecular genetic methods in the diagnosis of hematologic neoplasms. Semin Diagn Pathol. 2002;19:72–93. [PubMed] [Google Scholar]
- 6.Langerak AW, van Krieken JH, Wolvers-Tettero IL, et al. The role of molecular analysis of immunoglobulin and T cell receptor gene rearrangements in the diagnosis of lymphoproliferative disorders. J Clin Pathol. 2001;54:565–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gamadia LE, van Leeuwen EMM, Remmerswaal EBM, et al. The size and phenotype of virus-specific T cell populations is determined by repetitive antigenic stimulation and environmental cytokines. J Immunol Baltim Md. 19502004;172:6107–6114. [DOI] [PubMed] [Google Scholar]
- 8.Khan N, Shariff N, Cobbold M, et al. Cytomegalovirus seropositivity drives the CD8 T cell repertoire toward greater clonality in healthy elderly individuals. J Immunol Baltim Md. 19502002;169:1984–1992. [DOI] [PubMed] [Google Scholar]
- 9.Martin A, Barbesino G, Davies TF. T-cell receptors and autoimmune thyroid disease--signposts for T-cell-antigen driven diseases. Int Rev Immunol. 1999;18:111–140. [DOI] [PubMed] [Google Scholar]
- 10.Bristeau-Leprince A, Mateo V, Lim A, et al. Human TCR alpha/beta+ CD4-CD8- double-negative T cells in patients with autoimmune lymphoproliferative syndrome express restricted Vbeta TCR diversity and are clonally related to CD8+ T cells. J Immunol Baltim Md. 19502008;181:440–448. [DOI] [PubMed] [Google Scholar]
- 11.Ramesh M, Hamm D, Simchoni N, et al. Clonal and constricted T cell repertoire in common variable immune deficiency. Clin Immunol. 2015;doi: 10.1016/j.clim.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yew PY, Alachkar H, Yamaguchi R, et al. Quantitative characterization of T-cell repertoire in allogeneic hematopoietic stem cell transplant recipients. Bone Marrow Transplant. 2015;50:1227–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Naylor K, Li G, Vallejo AN, et al. The influence of age on T cell generation and TCR diversity. J Immunol Baltim Md. 19502005;174:7446–7452. [DOI] [PubMed] [Google Scholar]
- 14.Lazuardi L, Jenewein B, Wolf A, et al. Age-related loss of naïve T cells and dysregulation of T-cell/B-cell interactions in human lymph nodes. - PubMed - NCBI. Immunology. 2005;114:37–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Langerak AW, Groenen PJTA, Brüggemann M, et al. EuroClonality/BIOMED-2 guidelines for interpretation and reporting of Ig/TCR clonality testing in suspected lymphoproliferations. Leukemia. 2012;26:2159–2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Dongen JJM, Langerak AW, Brüggemann M, et al. Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: report of the BIOMED-2 Concerted Action BMH4-CT98-3936. Leukemia. 2003;17:2257–2317. [DOI] [PubMed] [Google Scholar]
- 17.Delfau MH, Hance AJ, Lecossier D, et al. Restricted diversity of V gamma 9-JP rearrangements in unstimulated human gamma/delta T lymphocytes. Eur J Immunol. 1992;22:2437–2443. [DOI] [PubMed] [Google Scholar]
- 18.van der Velden VHJ, Wijkhuijs JM, Jacobs DCH, et al. T cell receptor gamma gene rearrangements as targets for detection of minimal residual disease in acute lymphoblastic leukemia by real-time quantitative PCR analysis. Leukemia. 2002;16:1372–1380. [DOI] [PubMed] [Google Scholar]
- 19.Dippel E, Assaf C, Hummel M, et al. Clonal T-cell receptor gamma-chain gene rearrangement by PCR-based GeneScan analysis in advanced cutaneous T-cell lymphoma: a critical evaluation. J Pathol. 1999;188:146–154. [DOI] [PubMed] [Google Scholar]
- 20.Cushman-Vokoun AM, Connealy S, Greiner TC. Assay design affects the interpretation of T-cell receptor gamma gene rearrangements: comparison of the performance of a one-tube assay with the BIOMED-2-based TCRG gene clonality assay. J Mol Diagn. 2010;12:787–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lawnicki LC, Rubocki RJ, Chan WC, et al. The distribution of gene segments in T-cell receptor gamma gene rearrangements demonstrates the need for multiple primer sets. J Mol Diagn. 2003;5:82–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yakirevich E, Jackson CL, Meitner PA, et al. Analysis of T-cell clonality using laser capture microdissection and high-resolution microcapillary electrophoresis. J Mol Diagn. 2007;9:490–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Munro NJ, Snow K, Kant JA, et al. Molecular diagnostics on microfabricated electrophoretic devices: from slab gel- to capillary- to microchip-based assays for T- and B-cell lymphoproliferative disorders. Clin Chem. 1999;45:1906–1917. [PubMed] [Google Scholar]
- 24.Bruggemann M, White H, Gaulard P, et al. Powerful strategy for polymerase chain reaction-based clonality assessment in T-cell malignancies. Report of the BIOMED-2 Concerted Action BHM4 CT98-3936. Leukemia. 2007;21:215–222. [DOI] [PubMed] [Google Scholar]
- 25.Patel KP, Pan Q, Wang Y, et al. Comparison of BIOMED-2 versus laboratory-developed polymerase chain reaction assays for detecting T-cell receptor-gamma gene rearrangements. J Mol Diagn. 2010;12:226–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kansal R, Grody WW, Zhou J, et al. The value of T-Cell receptor (TRG) clonality evaluation by next-generation sequencing in clinical hematolymphoid tissues: a descriptive study of 41 cases from a single institution. Am J Clin Pathol. 2018;doi: 10.1093/ajcp/aqy046. [DOI] [PubMed] [Google Scholar]
- 27.Henegariu O, Heerema NA, Dlouhy SR, et al. Multiplex PCR: critical parameters and step-by-step protocol. BioTechniques. 1997;23:504–511. [DOI] [PubMed] [Google Scholar]
- 28.Altshuler M. Inadequate quality of ingredient. In: PCR Troubleshoooting: The Essential Guide. Norfolk, UK: Caister Academic Press; 2006: 20. [Google Scholar]
- 29.Derrieux C, Trinquand A, Bruneau J, et al. A single-tube, euroclonality-inspired, TRG clonality multiplex PCR Aids management of patients with enteropathic diseases, including from formaldehyde-fixed, paraffin-embedded tissues. J Mol Diagn. 2019;21:111–122. [DOI] [PubMed] [Google Scholar]
- 30.Dziubianau M, Hecht J, Kuchenbecker L, et al. TCR repertoire analysis by next generation sequencing allows complex differential diagnosis of T cell-related pathology. Am J Transplant. 2013;13:2842–2854. [DOI] [PubMed] [Google Scholar]
- 31.Schumacher JA, Duncavage EJ, Mosbruger TL, et al. A comparison of deep sequencing of TCRG rearrangements vs traditional capillary electrophoresis for assessment of clonality in T-Cell lymphoproliferative disorders. Am J Clin Pathol. 2014;141:348–359. [DOI] [PubMed] [Google Scholar]
- 32.Gong Q, Wang C, Zhang W, et al. Assessment of T-cell receptor repertoire and clonal expansion in peripheral T-cell lymphoma using RNA-seq data. Sci Rep. 2017;7:11301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Langerak AW, Molina TJ, Lavender FL, et al. Polymerase chain reaction-based clonality testing in tissue samples with reactive lymphoproliferations: usefulness and pitfalls: a report of the BIOMED-2 Concerted Action BMH4-CT98-3936. Leukemia. 2007;21:222–229. [DOI] [PubMed] [Google Scholar]
- 34.Greiner TC, Rubocki RJ. Effectiveness of capillary electrophoresis using fluorescent-labeled primers in detecting T-cell receptor gamma gene rearrangements. J Mol Diagn. 2002;4:137–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.