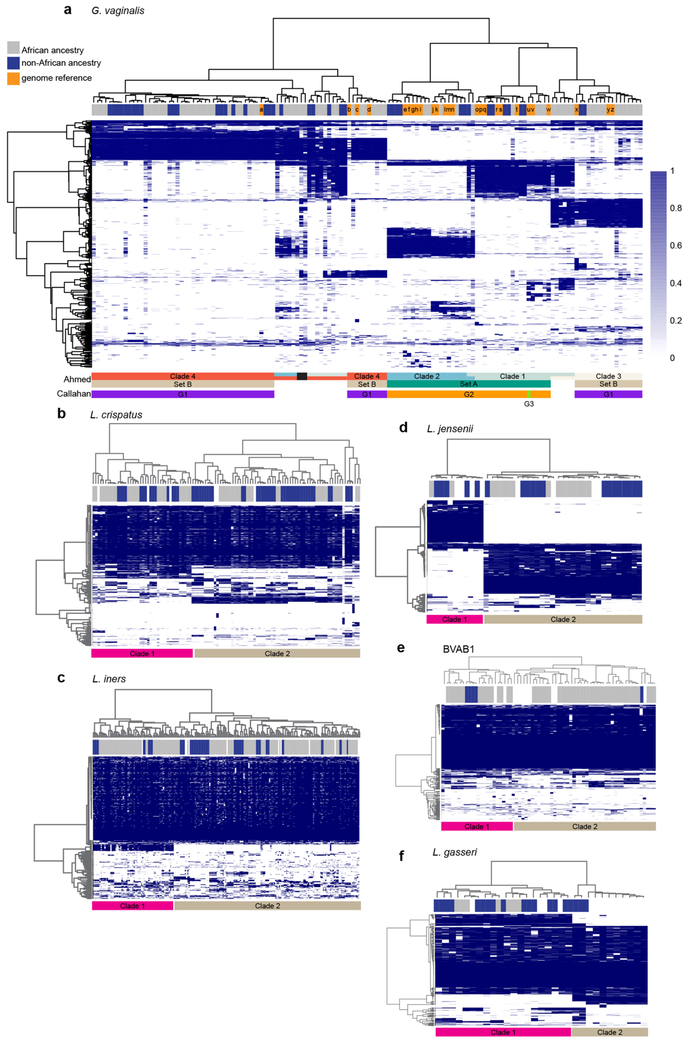
Extended Data Fig. 8 ∣. Association of G. vaginalis, L. crispatus, L. jensenii, L. gasseri, L. iners and Lachnospiracea BVAB1 strains with ancestry and other taxa.
Samples with these taxa were analysed in parallel with known reference strains using PanPhlan software to discriminate strain designations using default parameters of -min_coverage 1 (see Methods). a, G. vaginalis. Using these parameters, 121 samples provided sufficient numbers of G. vaginalis reads to provide accurate strain designations. Strain designations, which were previously reported by Ahmed et al.33 or Callahan et al.53, are indicated by the colored bars below the heat map. Note that G1 of Callahan et al. is within Set B of Ahmed et al., which also overlaps clades 3 and 4, and G2 of Callahan et al. includes Set A of Ahmed et al., which is also subdivided into clades 1 and 2. G3 of Callahan et al. classifies in clade 1 of Ahmed et al. The ancestry of each participant is indicated in the bar above the heat map, where blue indicates non-African and gray indicates African ancestry, and orange indicates a reference strain genome. Note: several samples contained multiple strains of different lineage. The black bar indicates two samples that contained three strains from clades 2, 3 and 4. b-f, L. crispatus, L. jensenii, L. gasseri, L. iners, and Lachnospiracea BVAB1. Analyses similar to that done for G. vaginalis above were performed with samples containing sufficient presence of these taxa (see above, and Methods). The ancestry of each participant is indicated in the bar above the heat map, where blue indicates non-African and gray indicates African ancestry, and white indicates a reference strain genome. Clades are differentiated by pink and light brown bars under each heat map.