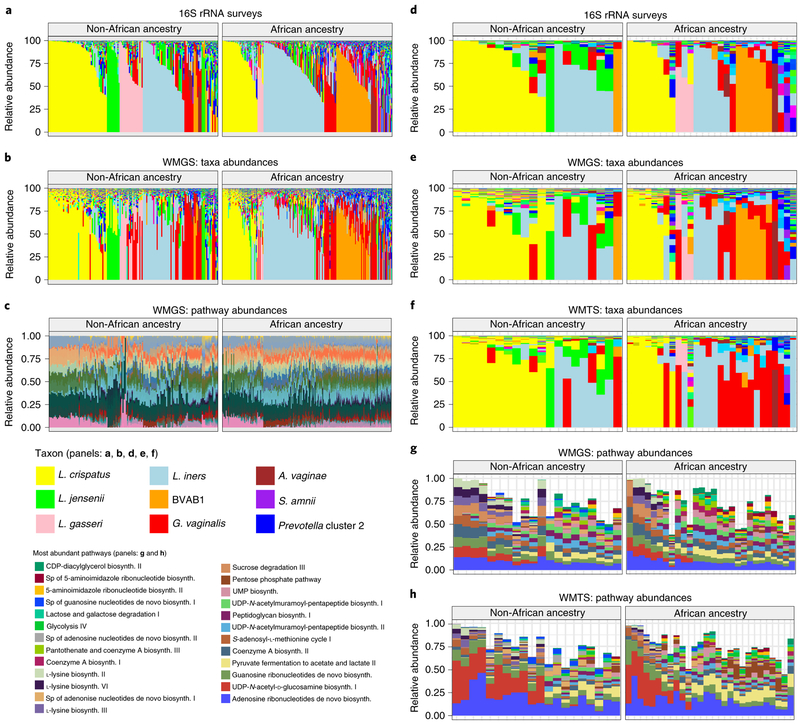
Fig. 5 ∣. Metagenomic, metatranscriptomic and pathway analyses of vaginal microbiome samples support metabolic differences among vagitypes of pregnant women of African and non-African ancestry.
Longitudinal samples from 90 women, 49 of African and 41 of non-African ancestry were compared. a, 16S rRNA taxonomic assignments of samples from all three trimesters from women of non-African and African ancestry. b, Taxonomic profiles from metagenomic sequence data of samples from all three trimesters from women of non-African and African ancestry. c, Relative abundances of metabolic pathways estimated using HUMAnN255 from metagenomic sequence data of samples from all three trimesters from women of non-African and African ancestry. d, 16S rRNA taxonomic assignments of samples from the second or early third trimester (one sample per pregnancy) from women of non-African and African ancestry. e, Taxonomic profiles from metagenomic sequence data from samples as in d. f, Taxonomic profiles from metatranscriptomic sequence from samples as in d. g, Relative abundances of 25 highly abundant metabolic pathways estimated using HUMAnN2 from metagenomic sequence data from samples as in d. h, Relative abundances of 25 highly abundant metabolic pathways estimated using HUMAnN2 from metatranscriptomic sequence from samples as in d. See Supplementary Table 6 for mapping statistics for b, e and f. ‘sp’ in the pathways key means ‘superpathway’.