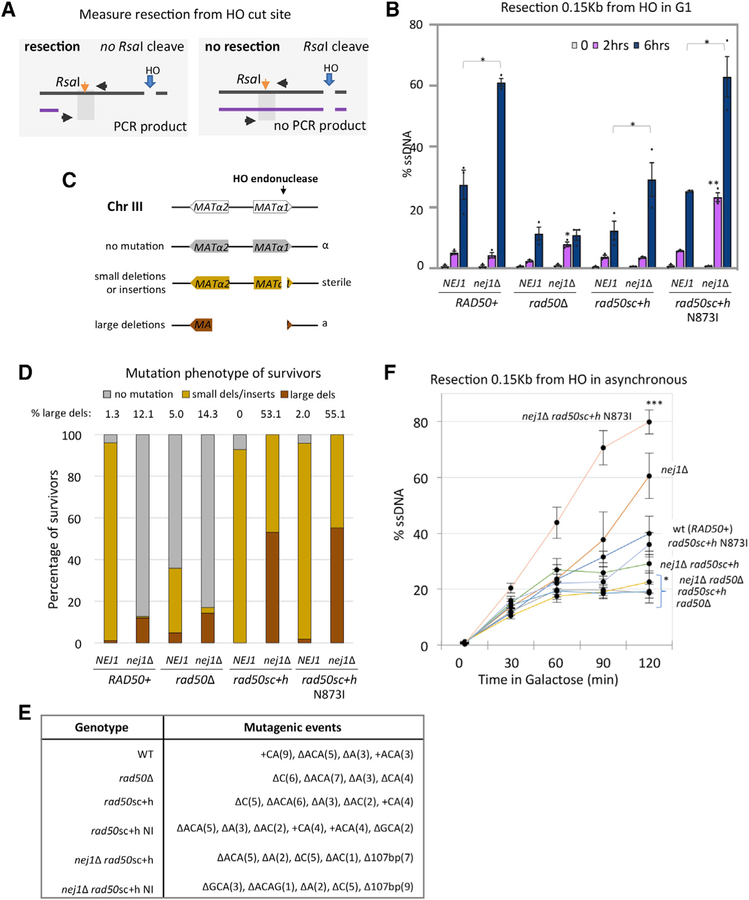
Figure 3. Interplay of DNA End-Tethering and Resection in Maintaining Genomic Stability.
(A) Schematic representation of regions around HO cut site on chromosome III. The RsaI site used in the qPCR resection assays is 0.15 kb from the DSB.
(B and F) Resection of DNA 0.15 kb away from the HO DSB, as measured by percent single-stranded DNA (ssDNA), at 0, 2, and 6 h post DSB induction in G1 cells (B) or at 0, 30, 60, 90, and 120 min post DSB induction in cycling cells (F) in WT (JC-3585), nej1Δ (JC-3884), rad50Δ (JC-3882), nej1Δ rad50Δ (JC-3887), rad50sc+h (JC-4458), nej1Δ rad50sc+h (JC-4471), rad50sc+h N873I (JC-4567), and nej1Δ rad50sc+h N873I (JC-4569). Analysis was performed in triplicate from at least three biological replicate experiments. Statistical analysis is described in STAR Methods.
(C) Schematic representation of mating-type analysis of survivors from persistent DSB induction assays. Disruption of the MATa1 gene results in a sterile phenotype, and disruption of the MATα2 gene (~700 bp upstream of HO cut site) results in an a-like phenotype in the mating type assays. The mating phenotype is a readout for the type of repair: α survivors (mutated HO endonuclease in gray), sterile survivors (small insertions or deletions in yellow), and a-like survivors (>700 bp deletion in red).
(D and E) The mating phenotype of survivors (D) and mutagenic events (E) were determined by DNA sequencing in 20 sterile survivors (except in nej1Δ and nej1Δ rad50Δ because few survived) and 20 a-like survivors (except WT, rad50sc+h, and rad50sc+h N873I) and shown in Figure S2. Strains include WT (JC-727), nej1Δ (JC-1342), rad50Δ (JC-3313), nej1Δ rad50Δ (JC-3314), rad50sc+h (JC-4424), nej1Δ rad50sc+h (JC-4476), rad50sc+h N873I (JC-4561), and nej1Δ rad50sc+h N873I (JC-4597).