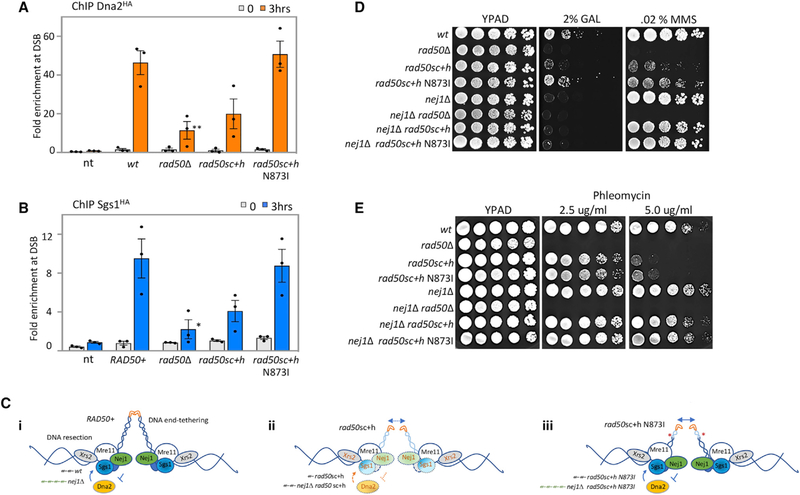
Figure 5. Role of Nej1 in Regulation of HR Repair Pathway.
(A) Enrichment of Dna2HA at DSB, at 0and 3-h time points, in WT (JC-4117), rad50Δ (JC-4503), rad50sc+h (JC-4531), rad50sc+h N873I (JC-4564), and no tag control (JC-727) were determined at 0.6 kb from DSB.
(B) Enrichment of Sgs1HA at DSB, at 0and 3-h time points, in WT (JC-4135), rad50Δ (JC-4138), rad50sc+h (JC-4457), rad50sc+h N873I (JC-4565), and no tag control (JC-727) were determined at 0.6 kb from DSB. The fold enrichment is normalized to recovery at the SMC2 locus. Analysis was performed in triplicate from at least three biological replicate experiments. Statistical analysis is described in STAR Methods.
(C) Model supported by microscopy data that determined tethering, qPCR that measured resection, and ChIP data assess the binding of Xrs2, Nej1, and Dna2Sgs1 at the DSB in (i) RAD50+, (ii) rad50sc+h, and (iii) rad50sc+h N873I.
(D and E) Drop assay with 5-fold serial dilutions on growth media YPAD, YPAD + 2% galactose, or YPAD + 0.02% MMS (D), and YPAD + 2.5 or 5 ug/ml phleomycin (E) using the following strains: WT (JC-727), rad50Δ (JC-3313), rad50sc+h (JC-4424), rad50sc+h N873I (JC-4561), nej1Δ (JC-1342), nej1Δ rad50Δ (JC-3314), nej1Δ rad50sc+h (JC-4476), and nej1Δ rad50sc+h N873I (JC-4597).