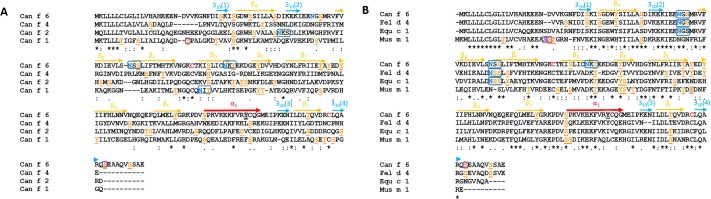
Fig 2. Primary structure alignment of lipocalins.
(A)The secondary structure for Can f 6 is drawn above the sequences. The colors correspond to the model of Can f 6 depicted in Fig 1 (A). Despite their conserved lipocalin architecture there is low sequence similarity between the dog lipocalins Can f 1, 2, 4, and Can f 6. The only disulfide bond (C, in red) occurs between C67 and C160. (B) Primary sequence alignment of the Can f 6 dog lipocalin, cat (Fel d 4), horse (Equ c 1), and mouse (Mus m 1). There is much more sequence similarity in these lipocalins compared to those of the dog lipocalins. Alignment residues are as: “*” Identical amino acid, “:” Similar amino acid, and “.” Slightly similar amino acid[35]. NxS/T (Blue N in blue boxes) are predicted N glycosylation sites (28), S/T (purple in black boxes) are predicted O glycosylation sites (29), S/T/T (orange) are predicted phosphorylation sites (30), and Y are predicted sulfation sites (31).