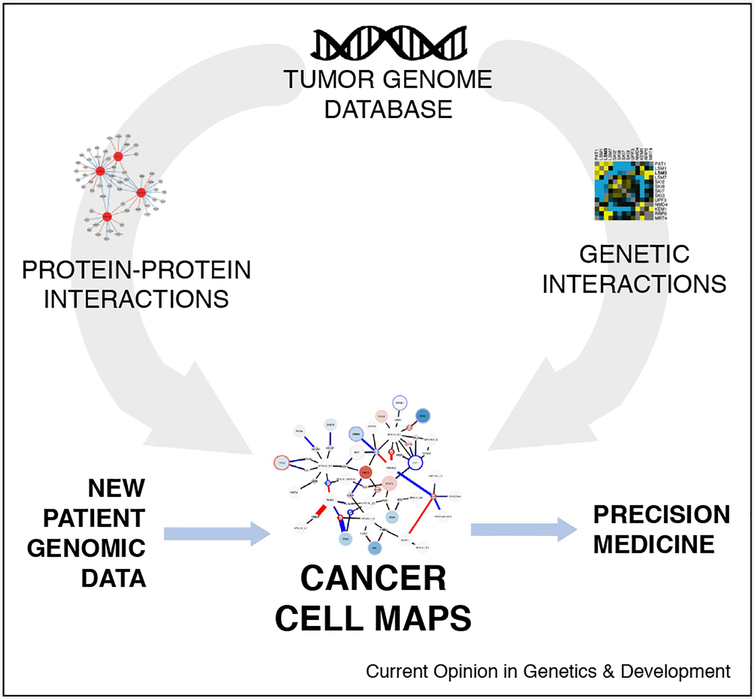
Figure 1.
The cancer cell map for precision oncology. The analysis of large numbers of tumor genomes has revealed key mutations that drive cancer pathology. However, connecting the genotype of tumor mutational profiles to phenotypes of drug sensitivity is impeded by a fundamental lack of understanding into how protein–protein (physical) and genetic (functional) interactions are configured to drive cellular processes, such as proliferation and death. Seeded by this large-scale analysis of tumor genomes (top), what is needed is a systematic, context-diverse network mapping of the protein–protein interactions for mutated and wild-type oncogenes and tumor suppressors (left) as well as the identification of synthetic lethal genetic interactions (right). PPI and GI mapping, two highly complementary data sources, can then be assembled to form Cancer Cell Maps, providing a working scaffold of molecular interactions and the cell types/conditions under which they are active. New patient data can then be queried against this resource by mapping alterations to these hallmark cancer networks, which can inform precision medicine (blue arrows).