Figure 3.
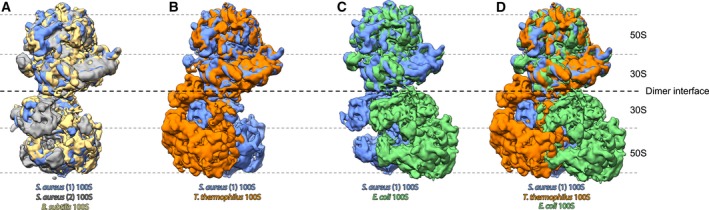
Conformations of 100S structures from different bacteria. Superposition of (A) Bacillus subtilis 100S (EMD‐3664, yellow) and two independently studied Staphylococcus aureus 100S (EMD‐3637, gray and EMD‐3638, blue) structures; (B) Thermus thermophilus 100S (colored orange) and S. aureus 100S (EMD‐3638, blue) structures; (C) Escherichia coli 100S (EMD‐0139, green) and S. aureus 100S (EMD‐3638, blue) structures; and (D) T. thermophilus 100S, E. coli 100S and S. aureus 100S (same coloring as A–C) structures. Both S. aureus 100S and B. subtilis 100S structures show a high degree of conformational conservation while T. thermophilus and E. coli 100S ribosomes are clearly staggered in a different conformation (thus, in B‐D, only one of the S. aureus 100S is shown as a representative of the second S. aureus 100S as well as B. subtilis 100S).
