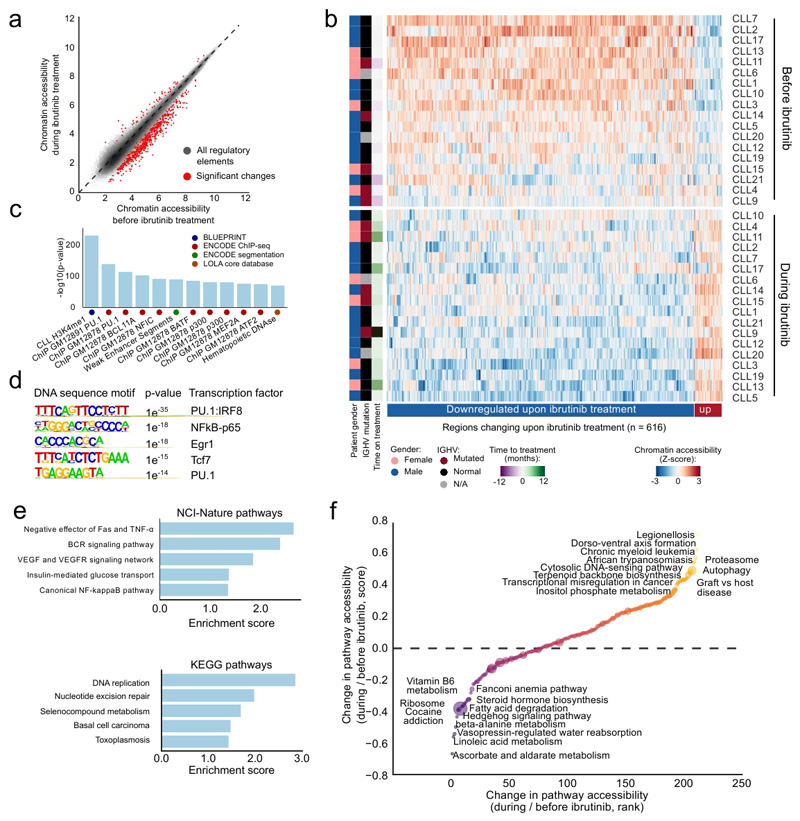
Figure 3. Differential analysis of ibrutinib-induced changes in chromatin accessibility for matched CLL patient samples.
(a) Scatterplot comparing ATAC-seq signal intensities across all open chromatin sites between samples collected before and during ibrutinib treatment. Significant changes correspond to an FDR-adjusted p-value below 0.01 and an absolute log2 fold change above 1 as calculated by the DESeq2 software. The diagonal is shown as a dashed line, as a reference indicating regions with no change in chromatin accessibility upon ibrutinib treatment. (b) Heatmap of normalized chromatin accessibility (Z-scores) for all regions with significantly differential chromatin accessibility according to panel a. (c) Region set enrichment analysis for genomic regions with reduced chromatin accessibility during ibrutinib treatment based on LOLA analysis, showing the twelve most significantly enriched region sets. (d) De novo motif enrichment analysis for regions with reduced chromatin accessibility during ibrutinib treatment. Reported p-values were calculated by the HOMER software using a binomial test. (e) Gene set analysis for enrichment of NCI-Nature and KEGG pathways among genes located in the vicinity of regions with reduced chromatin accessibility during ibrutinib treatment. Enrichment scores were calculated by the Enrichr software and represent the log p-value of a Fisher’s exact test multiplied by a Z-score of deviation from the expected rank. (f) Pathway-centric assessment of changes in chromatin accessibility induced by ibrutinib treatment. Normalized ATAC-seq signals of all genes in each KEGG pathway were aggregated to rank pathways. Yellow/orange dots denote pathways characterized by higher chromatin accessibility during ibrutinib treatment than before ibrutinib treatment, while blue/purple dots indicate pathways with lower chromatin accessibility.