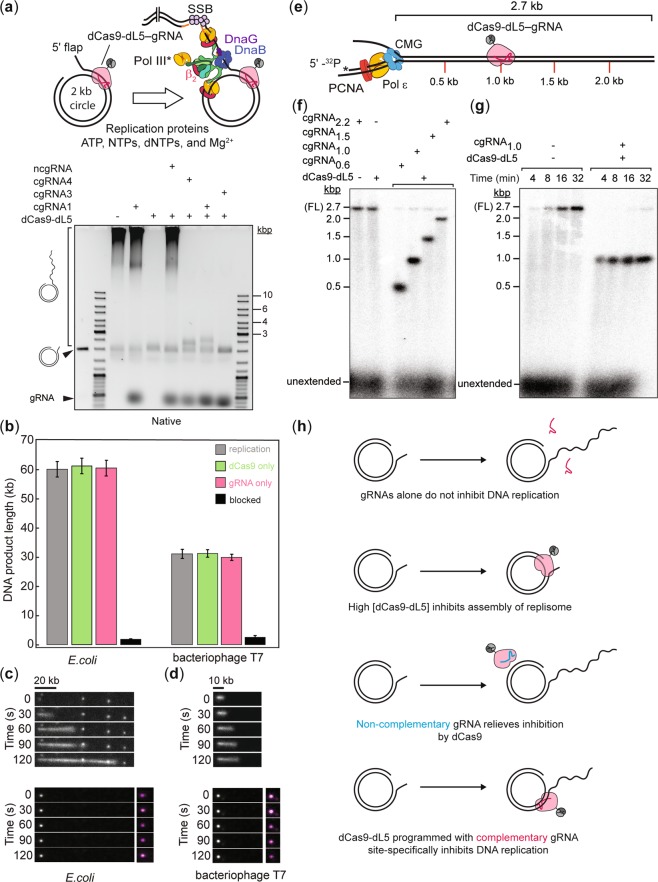
Figure 2.
dCas9-dL5 efficiently and stably blocks bacterial, viral, and eukaryotic DNA replication regardless of the targeted strand. (a) Schematic of the leading-lagging rolling-circle DNA replication assay. Addition of the E. coli or T7 replication proteins, nucleotides, and Mg2+ initiates DNA synthesis in a leading strand replication reaction. The DNA products are separated by gel electrophoresis and visualized by staining with SYBR-Gold, or visualized by single-molecule fluorescence microscopy by staining with Sytox orange. dCas9-dL5 (100 nM) programmed with ncgRNA (400 nM) and cgRNAs alone (400 nM) alone do not inhibit DNA replication. At high concentrations, dCas9-dL5 (100 nM) alone inhibits DNA synthesis. dCas9-dL5 programmed with cgRNAs arrest the progress of the replication fork at the target site. See also, Fig. S2 (b) Bar plots of mean DNA product lengths from E. coli and T7 single-molecule rolling-circle DNA replication assays. Values plotted are derived from exponential fits to single-molecule DNA product length distributions (n > 91 molecules). Error bars indicate errors of the fit. (c,d) (Top panel) Example kymographs of an individual DNA molecule undergoing DNA replication by E. coli (c) (n = 177 molecules; replication efficiency of 26 ± 2% (SEM)) and T7 replisomes (d) (n = 136 molecules; replication efficiency of 24 ± 2% (SEM)) in the absence of target bound dCas9-dL5. (Bottom panels) Example kymographs of an individual DNA molecule arrested by target bound dCas9-dL5. The grey scale indicates the fluorescence intensity of stained DNA and magenta indicates dCas9-dL5–cgRNA stained by MGE. No replication events were detected. (e) Schematic of the eukaryotic DNA replication assay. Eukaryotic replication is blocked by dCas9-dL5 at specific positions on the replication template. (f) dCas9-dL5 efficiently blocks eukaryotic replication. The cgRNAs used to specifically target the template are indicated. cgRNA0.6 and cgRNA2.2 block the leading strand; cgRNA1.0 and cgRNA1.5 block the lagging strand (see Supplementary Methods for details). All reactions were stopped at 16 min. (g) Time course of eukaryotic replication in the presence or absence of dCas9-dL5 and cgRNA1.0. Reactions were stopped at 4, 8, 16 and 32 min as indicated. (h) Summary of interactions of dCas9-dL5 and template DNA. Only the correctly programmed dCas9-dL5-cgRNA complex site specifically inhibits DNA replication.