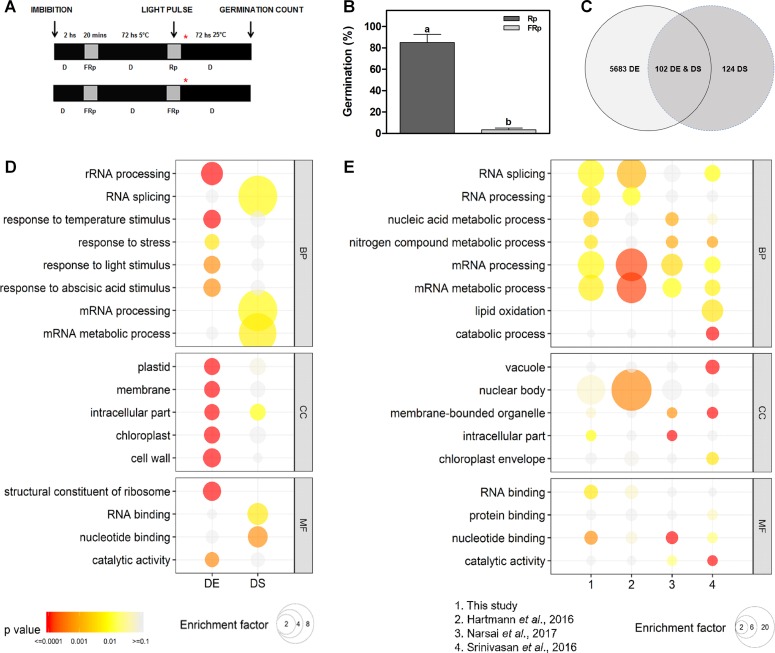
Figure 1.
Germination induced by a red pulse modifies the alternative splicing pattern of 226 genes in Col-0 seeds. (A) Scheme of the experimental protocol. Col-0 seeds were irradiated with an Rp or an FRp after 3 days of chilling, and samples for RNA isolation were collected 12 h after the light pulse. Asterisk shows sampling point for RNA extractions. (B) Germination percentage of Col-0 seeds irradiated with an Rp (85%) or an FRp. Each bar represents mean ± SE (n = 3). Significant differences between means are shown by different letters (p < 0.05 by Student’s t-test). (C) Overlap between genes differentially affected by light at the expression level (DE genes) and genes regulated by light at the AS level (DS genes). (D) GO enrichment analysis comparing the DE and DS genes of our study. (E) GO enrichment analysis comparing DS genes between our study, and the works by Hartmann et al. (2016), Narsai et al. (2017), and Srinivasan et al. (2016). For panels D and E, GO was evaluated at three different levels: biological processes (BP), cellular component (CC), and molecular function (MF). The color gradient represents adjusted p-values, and the differences in bubble size correlate with the enrichment factor. Only those categories showing a statistically significant enrichment at either gene expression or AS level are depicted. AS, alternative splicing; Col-0, Columbia-0; DE, differentially expressed; DS, differentially spliced; FRp, far-red pulse; GO, gene ontology; Rp, red pulse.