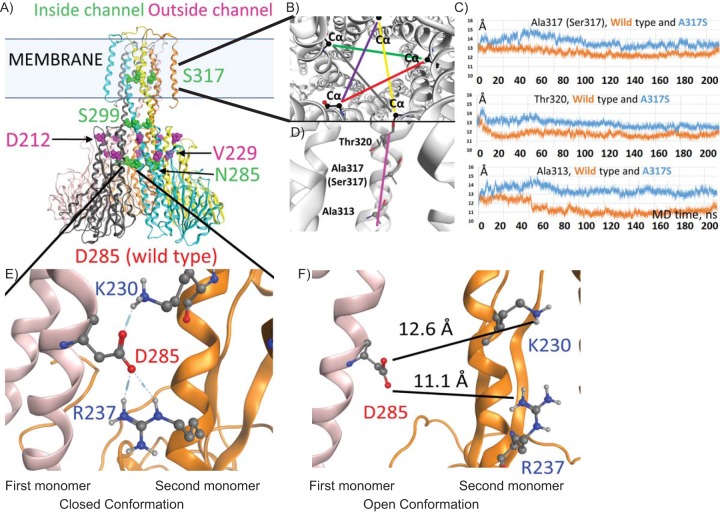
FIG 3.
Potential mechanism of CorA-mediated resistance to 4HQs. (A) Model of M. tuberculosis CorA based on T. maritima crystal structures and Molecular Operating Environment (MOE) homology program prediction of the structural location of amino acid residues affected by corA mutations in the M. tuberculosis-resistant clones. (B) Changes in the channel diameter around the mutated residue. Five Cα distances that are related to the channel diameter are shown in different colors (as viewed from the periplasmic side, top-down). (C) Plots showing the average value for the five distances (in panel B) taken at each 20-ps MD trajectory frame. (D) The part of the helix facing the channel (magenta) shows A317S and neighboring residues Thr320 and Ala313. (E) The predicted H-bonding between D285 and the adjoining monomer in the closed conformation. (F) Model of the D285 mutation of M. tuberculosis CorA using the open conformation PDB 3jcg from the TmCorA structure, predicting an increase in distance between D285 and the neighboring monomer.