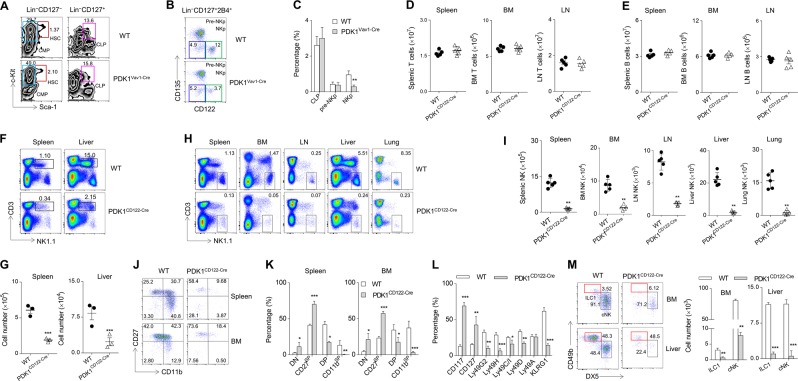
Fig. 1.
CD122-Cre mediated pdk1 deletion severely compromises NK cells development. a-c Representative flow cytometry plots (a, b) and quantification (c) of hematopoietic stem cells (HSC, Lin−CD127−c-Kit+Sca-1+), common myeloid progenitors (CMP, Lin−CD127−c-Kit+Sca-1−) and common lymphoid progenitors (CLP, Lin−CD127+c-Kit+Sca-1+) (A), pre-NKp (Lin−CD127+2B4+CD135−CD112−) and NKp (Lin−CD127+2B4+CD135−CD112+) (b) in the BM of WT and PDK1Vav1-Cre mice. Numbers near the indicated square box show the respective percentage. d, e The absolute number of T cells (d) and B cells (e) in the indicated tissues and organs from WT and PDK1CD122-Cre mice. f, g Representative flow cytometric profiles (f) and the absolute number (g) of NK-T cells (CD3lowNK1.1low) in the spleens and livers of WT and PDK1CD122-Cre mice. h, i Representative flow cytometric profiles (h) and the absolute number (i) of NK cells (CD3−NK1.1+) in the spleen, BM, LN, liver, and lungs of WT and PDK1CD122-Cre mice. j, k Representative flow cytometric profiles (j) and the percentages (k) of NK cell subsets in the spleen and BM from the WT and PDK1CD122-Cre mice. DN (CD27−CD11b−), CD27 SP (CD27+CD11b-), DP (CD27+CD11b+) and CD11b SP (CD27-CD11b+) cells. l Percentage of developmental markers on splenic NK cells (CD3−NK1.1+) in WT and PDK1CD122-Cre mice. m Representative flow cytometry plots and quantification of BM and liver ILC1 (CD3−NK1.1+CD49a+CD49b−). The data represent one of three independent experiments, and values are expressed as the mean ± s.d